4U9G
 
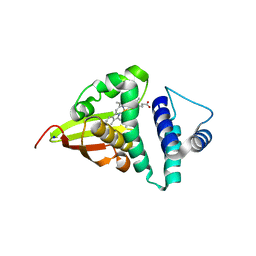 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)CO ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | CARBON MONOXIDE, NO-binding heme-dependent sensor protein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9J
 
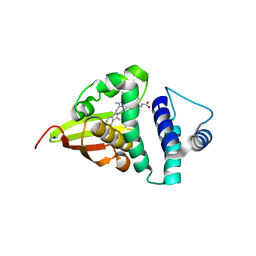 | | Crystal structure of an H-NOX protein from S. oneidensis in the Mn(II) ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, NO-binding heme-dependent sensor protein, SODIUM ION, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9B
 
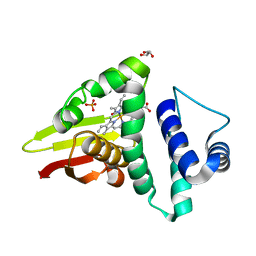 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)NO ligation state | | Descriptor: | GLYCEROL, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9K
 
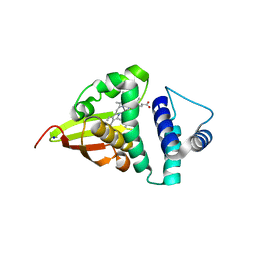 | | Crystal structure of an H-NOX protein from S. oneidensis in the Mn(II)NO ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U99
 
 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II) ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | NO-binding heme-dependent sensor protein, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4IT2
 
 | | Mn(III)-PPIX bound Tt H-NOX | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, Methyl-accepting chemotaxis protein | | Authors: | Winter, M.B, Klemm, P.J, Phillips-Piro, C.M, Raymond, K.M, Marletta, M.A. | | Deposit date: | 2013-01-17 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Porphyrin-Substituted H-NOX Proteins as High-Relaxivity MRI Contrast Agents.
Inorg.Chem., 52, 2013
|
|
3LAI
 
 | | Structural insights into the molecular mechanism of H-NOX activation | | Descriptor: | IMIDAZOLE, Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Olea Jr, C, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Structural insights into the molecular mechanism of H-NOX activation.
Protein Sci., 19, 2010
|
|
3LAH
 
 | | Structural insights into the molecular mechanism of H-NOX activation | | Descriptor: | IMIDAZOLE, Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Olea Jr, C, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the molecular mechanism of H-NOX activation.
Protein Sci., 19, 2010
|
|
3MVC
 
 | | High resolution crystal structure of the heme domain of GLB-6 from C. elegans | | Descriptor: | Globin protein 6, NITRATE ION, PRASEODYMIUM ION, ... | | Authors: | Yoon, J, Herzik Jr, M.A, Winter, M.B, Tran, R, Olea Jr, C, Marletta, M.A. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structure and properties of a bis-histidyl ligated globin from Caenorhabditis elegans.
Biochemistry, 49, 2010
|
|
3IQB
 
 | | Tt I75F/L144F H-NOX | | Descriptor: | Methyl-accepting chemotaxis protein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Weinert, E.E, Plate, L, Whited, C.A, Olea Jr, C, Marletta, M.A. | | Deposit date: | 2009-08-19 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Determinants of Ligand Affinity and Heme Reactivity in H-NOX Domains.
Angew.Chem.Int.Ed.Engl., 49, 2009
|
|
3TF1
 
 | | Crystal structure of an H-NOX protein from T. tengcongensis under 6 atm of xenon | | Descriptor: | ACETATE ION, Methyl-accepting chemotaxis protein, OXYGEN MOLECULE, ... | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0369 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TFF
 
 | | Crystal structure of an H-NOX protein from Nostoc sp. PCC 7120, L67W mutant | | Descriptor: | Alr2278 protein, MALONIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9401 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TF0
 
 | | Crystal structure of an H-NOX protein from T. tengcongensis | | Descriptor: | ACETATE ION, Methyl-accepting chemotaxis protein, OXYGEN MOLECULE, ... | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TFE
 
 | | Crystal structure of an H-NOX protein from Nostoc sp. PCC 7120, L66W mutant under 6 atm of xenon | | Descriptor: | Alr2278 protein, MALONIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6BDE
 
 | | Crystal structure of Fe(II) unliganded H-NOX protein mutant A71G from K. algicida | | Descriptor: | 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bruegger, J.J, Hespen, C.W, Marletta, M.A. | | Deposit date: | 2017-10-22 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Native Alanine Substitution in the Glycine Hinge Modulates Conformational Flexibility of Heme Nitric Oxide/Oxygen (H-NOX) Sensing Proteins.
ACS Chem. Biol., 13, 2018
|
|
6BDD
 
 | | Crystal structure of Fe(II) unliganded H-NOX protein from K. algicida | | Descriptor: | 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bruegger, J.J, Hespen, C.W, Marletta, M.A. | | Deposit date: | 2017-10-22 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Native Alanine Substitution in the Glycine Hinge Modulates Conformational Flexibility of Heme Nitric Oxide/Oxygen (H-NOX) Sensing Proteins.
ACS Chem. Biol., 13, 2018
|
|
5UFV
 
 | |
3TFD
 
 | | Crystal structure of an H-NOX protein from Nostoc sp. PCC 7120, L66W mutant | | Descriptor: | Alr2278 protein, MALONIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TFG
 
 | | Crystal structure of an H-NOX protein from Nostoc sp. PCC 7120, L66W/L67W double mutant | | Descriptor: | Alr2278 protein, MALONIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9003 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TFA
 
 | | Crystal structure of an H-NOX protein from Nostoc sp. PCC 7120 under 6 atm of xenon | | Descriptor: | Alr2278 protein, PROTOPORPHYRIN IX CONTAINING FE, XENON | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2711 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TF8
 
 | | Crystal structure of an H-NOX protein from Nostoc sp. PCC 7120 | | Descriptor: | Alr2278 protein, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1302 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TF9
 
 | | Crystal structure of an H-NOX protein from Nostoc sp. PCC 7120 under 1 atm of xenon | | Descriptor: | Alr2278 protein, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5901 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3EEE
 
 | | Probing the function of heme distortion in the H-NOX family | | Descriptor: | CHLORIDE ION, Methyl-accepting chemotaxis protein, OXYGEN MOLECULE, ... | | Authors: | Olea Jr, C, Boon, E.M, Pellicena, P, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2008-09-04 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Probing the function of heme distortion in the H-NOX family.
Acs Chem.Biol., 3, 2008
|
|
3ET6
 
 | | The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase | | Descriptor: | PHOSPHATE ION, Soluble guanylyl cyclase beta | | Authors: | Winger, J.A, Derbyshire, E.R, Lamers, M.H, Marletta, M.A, Kuriyan, J. | | Deposit date: | 2008-10-07 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase.
Bmc Struct.Biol., 8, 2008
|
|
6PAS
 
 | | Inactive State of Manduca sexta soluble guanylate cyclase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Soluble guanylyl cyclase alpha-1 subunit, Soluble guanylyl cyclase beta-1 subunit | | Authors: | Yokom, A.L, Horst, B.G, Marletta, M.A, Hurley, J.H. | | Deposit date: | 2019-06-11 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy.
Elife, 8, 2019
|
|
