1Y8R
 
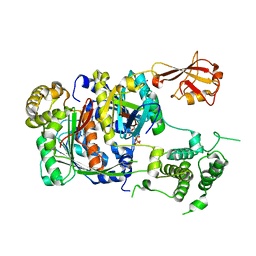 | | SUMO E1 ACTIVATING ENZYME SAE1-SAE2-SUMO1-MG-ATP COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ubiquitin-like 1 activating enzyme E1A, ... | | Authors: | Lois, L.M, Lima, C.D. | | Deposit date: | 2004-12-13 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1
Embo J., 24, 2005
|
|
1Y8Q
 
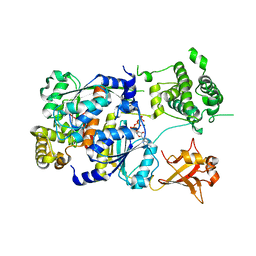 | | SUMO E1 ACTIVATING ENZYME SAE1-SAE2-MG-ATP COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ubiquitin-like 1 activating enzyme E1A, ... | | Authors: | Lois, L.M, Lima, C.D. | | Deposit date: | 2004-12-13 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1
Embo J., 24, 2005
|
|
6GUM
 
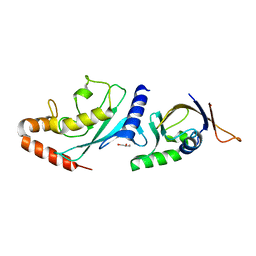 | | Structure of the A.thaliana E1 UFD domain in complex with E2 | | Descriptor: | GLYCEROL, SAE2, SUMO-conjugating enzyme SCE1 | | Authors: | Liu, B, Lois, L.M, Reverter, D. | | Deposit date: | 2018-06-19 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural insights into SUMO E1-E2 interactions in Arabidopsis uncovers a distinctive platform for securing SUMO conjugation specificity across evolution.
Biochem.J., 476, 2019
|
|
6GV3
 
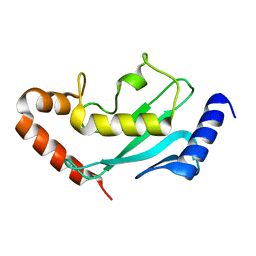 | |
8OI3
 
 | | Structure of NopD with AtSUMO2 | | Descriptor: | Small ubiquitin-related modifier 2, Type III effector, prop-2-en-1-amine | | Authors: | Reverter, D, Li, Y. | | Deposit date: | 2023-03-22 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Broad-spectrum ubiquitin/ubiquitin-like deconjugation activity of the rhizobial effector NopD from Bradyrhizobium (sp. XS1150).
Commun Biol, 7, 2024
|
|
2OIV
 
 | | Structural Analysis of Xanthomonas XopD Provides Insights Into Substrate Specificity of Ubiquitin-like Protein Proteases | | Descriptor: | PHOSPHATE ION, Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
2OIX
 
 | | Xanthomonas XopD C470A Mutant | | Descriptor: | Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
