6CC2
 
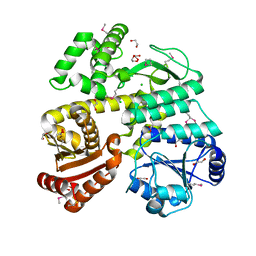 | | Crystal Structure of CDC45 from Entamoeba histolytica | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cell division control protein 45 cdc45 putative, ... | | Authors: | Shi, K, Kurniawan, F, Kurahashi, K, Bielinsky, A, Aihara, H. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure ofEntamoeba histolyticaCdc45 Suggests a Conformational Switch that May Regulate DNA Replication.
iScience, 3, 2018
|
|
6XC1
 
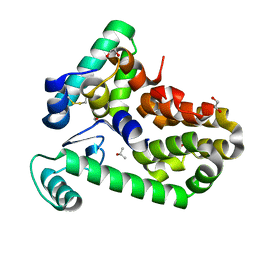 | | Crystal structure of bacteriophage T4 spackle and lysozyme in orthorhombic form | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Lysozyme, ... | | Authors: | Shi, K, Oakland, J.T, Kurniawan, F, Moeller, N.H, Aihara, H. | | Deposit date: | 2020-06-07 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of superinfection exclusion by bacteriophage T4 Spackle.
Commun Biol, 3, 2020
|
|
6XC0
 
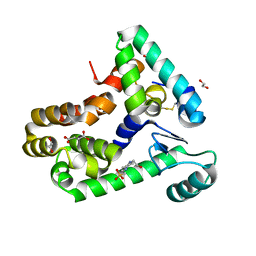 | | Crystal structure of bacteriophage T4 spackle and lysozyme in monoclinic form | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Shi, K, Oakland, J.T, Kurniawan, F, Moeller, N.H, Aihara, H. | | Deposit date: | 2020-06-07 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis of superinfection exclusion by bacteriophage T4 Spackle.
Commun Biol, 3, 2020
|
|
6X6O
 
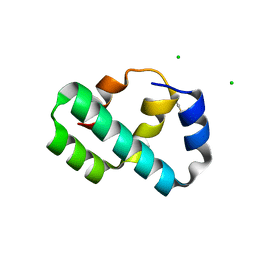 | | Crystal structure of T4 protein Spackle as determined by native SAD phasing | | Descriptor: | CHLORIDE ION, Protein spackle | | Authors: | Shi, K, Kurniawan, F, Banerjee, S, Moeller, N.H, Aihara, H. | | Deposit date: | 2020-05-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of bacteriophage T4 Spackle as determined by native SAD phasing.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VOY
 
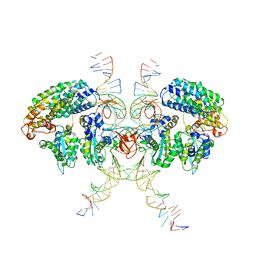 | | Cryo-EM structure of HTLV-1 instasome | | Descriptor: | DNA (25-MER), DNA (5'-D(P*AP*CP*AP*CP*AP*CP*TP*TP*GP*AP*CP*TP*AP*GP*GP*GP*TP*G)-3'), DNA-binding protein 7d, ... | | Authors: | Bhatt, V, Shi, K, Sundborger, A, Aihara, H. | | Deposit date: | 2020-02-01 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of host protein hijacking in human T-cell leukemia virus integration.
Nat Commun, 11, 2020
|
|
5WFY
 
 | |
7KM6
 
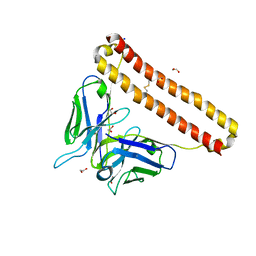 | | APOBEC3B antibody 5G7 Fv-clasp | | Descriptor: | 1,2-ETHANEDIOL, 5G7 human monoclonal FAB heavy chain, 5G7 human monoclonal FAB light chain, ... | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2020-11-02 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Characterization of a Minimal Antibody against Human APOBEC3B.
Viruses, 13, 2021
|
|
6DRT
 
 | |
6DT1
 
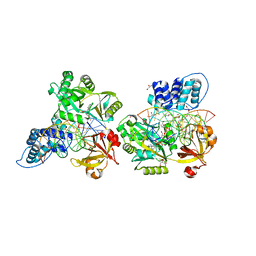 | | Crystal structure of the ligase from bacteriophage T4 complexed with DNA intermediate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | T4 DNA ligase structure reveals a prototypical ATP-dependent ligase with a unique mode of sliding clamp interaction.
Nucleic Acids Res., 46, 2018
|
|
