5BUM
 
 | | Crystal Structure of LysM domain from Equisetum arvense chitinase A | | Descriptor: | Chitinase A, SULFATE ION | | Authors: | Kitaoku, Y, Numata, T, Ohnuma, T, Taira, T, Fukamizo, T. | | Deposit date: | 2015-06-04 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure, mechanism, and phylogeny of LysM-chitinase conjugates specifically found in fern plants.
Plant Sci., 321, 2022
|
|
5K2L
 
 | | Crystal structure of LysM domain from Volvox carteri chitinase | | Descriptor: | 1,2-ETHANEDIOL, Chitinase, lysozyme | | Authors: | Kitaoku, Y, Numata, T, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chitin oligosaccharide binding to the lysin motif of a novel type of chitinase from the multicellular green alga, Volvox carteri.
Plant Mol. Biol., 93, 2017
|
|
7EBI
 
 | | Chitin-specific solute binding protein from Vibrio harveyi co-crystalized with chitotetraose. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
7EBM
 
 | | W363A mutant of Chitin-specific solute binding protein from Vibrio harveyi in complex with chitobiose. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2021-03-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
6LZU
 
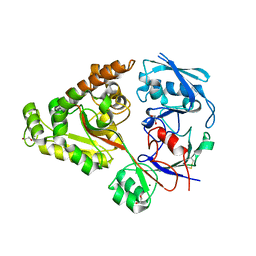 | | F411A mutant of chitin-specific solute binding protein from Vibrio harveyi co-crystalized with chitobiose. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2020-02-19 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
6LZV
 
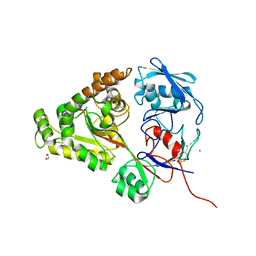 | | F437A mutant of chitin-specific solute binding protein from Vibrio harveyi co-crystalized with chitobiose. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2020-02-19 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
6LZW
 
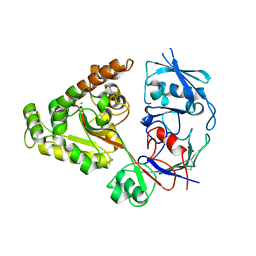 | | W513A mutant of chitin-specific solute binding protein from Vibrio harveyi co-crystalized with chitobiose. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2020-02-19 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
6LZT
 
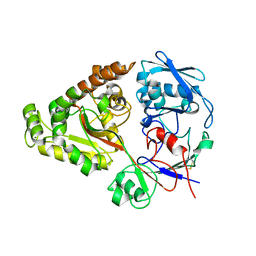 | | N409A mutant of chitin-specific solute binding protein from Vibrio harveyi co-crystalized with chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2020-02-19 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
6LZQ
 
 | | Chitin-specific solute binding protein from Vibrio harveyi in complex with chitotriose. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2020-02-19 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
5YZ6
 
 | | Solution structure of LysM domain from a chitinase derived from Volvox carteri | | Descriptor: | Chitinase, lysozyme | | Authors: | Kitaoku, Y, Nishimura, S, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2017-12-13 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structures and chitin-binding properties of two N-terminal lysin motifs (LysMs) found in a chitinase from Volvox carteri.
Glycobiology, 29, 2019
|
|
5YZK
 
 | | Solution structure of LysM domain from a chitinase derived from Volvox carteri | | Descriptor: | Chitinase, lysozyme | | Authors: | Kitaoku, Y, Nishimura, S, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structures and chitin-binding properties of two N-terminal lysin motifs (LysMs) found in a chitinase from Volvox carteri.
Glycobiology, 29, 2019
|
|
5YLG
 
 | | Crystal Structure of LysM domain from pteris ryukyuensis chitinase A | | Descriptor: | 1,2-ETHANEDIOL, Chitinase A, ZINC ION | | Authors: | Ohnuma, T, Kitaoku, Y, Umemoto, N, Numata, T, Fukamizo, T. | | Deposit date: | 2017-10-17 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of LysM domain from pteris ryukyuensis chitinase A
To Be Published
|
|
4ZXE
 
 | | X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/Chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZY9
 
 | | X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | Glucanase/chitosanase | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-21 | | Release date: | 2016-04-13 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZZ8
 
 | | X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Glucanase/chitosanase, ... | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZZ5
 
 | | X-ray crystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
5YQW
 
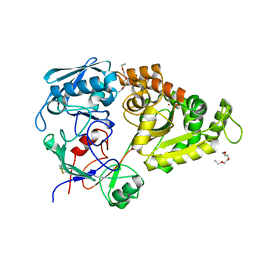 | | Structure and function of a novel periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICKEL (II) ION, ... | | Authors: | Suginta, W, Sritho, N, Ranok, A, Kitaoku, Y, Bulmer, D.M, van den Berg, B, Fukamizo, T. | | Deposit date: | 2017-11-08 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and function of a novel periplasmic chitooligosaccharide-binding protein from marineVibriobacteria.
J. Biol. Chem., 293, 2018
|
|
4RL3
 
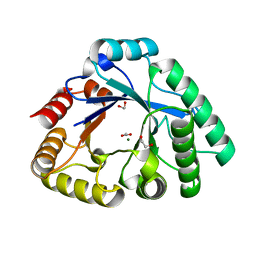 | | Crystal Structure of the Catalytic Domain of a family GH18 Chitinase from fern, Peteris ryukyuensis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Chitinase A, ... | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A class III chitinase without disulfide bonds from the fern, Pteris ryukyuensis: crystal structure and ligand-binding studies.
Planta, 242, 2015
|
|
6SGF
 
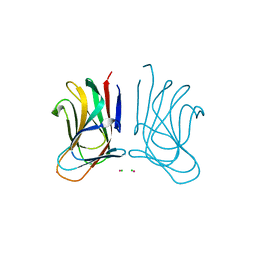 | | Molecular insight into a new low affinity xylan binding module CBM86, from the xylanolytic gut symbiont Roseburia intestinalis. | | Descriptor: | Beta-xylanase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Ejby, M, Abou Hachem, M, Leth, M.L, Guskov, A, Slotboom, D. | | Deposit date: | 2019-08-04 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Molecular insight into a new low-affinity xylan binding module from the xylanolytic gut symbiont Roseburia intestinalis.
Febs J., 287, 2020
|
|
2RV9
 
 | |
2RVA
 
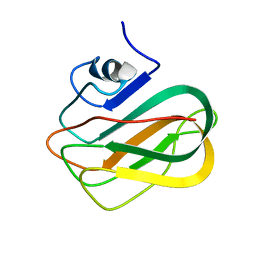 | |
7X56
 
 | | A CBg-ParM filament with ADP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ParM/StbA family protein | | Authors: | Koh, A, Ali, S, Robinson, R, Narita, A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A new actin-like filament from Clostridium botulinum exhibits a new system of filament turn over
To Be Published
|
|
7X54
 
 | | A CBg-ParM filament with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ParM/StbA family protein | | Authors: | Koh, A, Ali, S, Robinson, R, Narita, A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A new actin-like filament from Clostridium botulinum exhibits a new system of filament turn over
To Be Published
|
|
7X59
 
 | | A CBg-ParM filament with GTP or GDPPi | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, ParM/StbA family protein | | Authors: | Koh, A, Ali, S, Robinson, R, Narita, A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | A new actin-like filament from Clostridium botulinum exhibits a new system of filament turn over
To Be Published
|
|
7X55
 
 | |
