4LMY
 
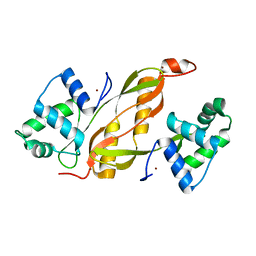 | | Structure of GAS PerR-Zn-Zn | | Descriptor: | Peroxide stress regulator PerR, FUR family, ZINC ION | | Authors: | Lin, C.S, Chao, S.Y, Nix, J.C, Tseng, H.L, Tsou, C.C, Fei, C.H, Ciou, H.S, Jeng, U.S, Lin, Y.S, Chuang, W.J, Wu, J.J, Wang, S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinct structural features of the peroxide response regulator from group a streptococcus drive DNA binding
Plos One, 9, 2014
|
|
6KL5
 
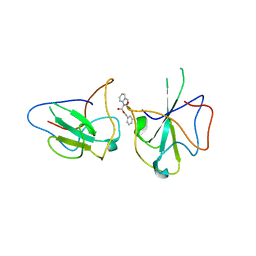 | | Structure of The N-terminal domain of Middle East respiratory syndrome coronavirus Nucleocapsid Protein complexed with Benzyl 2-(Hydroxymethyl)-1-Indolinecarboxylate | | Descriptor: | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Hsu, J.N, Wang, Y.S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
7WVH
 
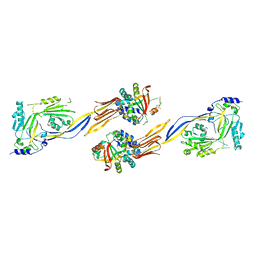 | |
7DYD
 
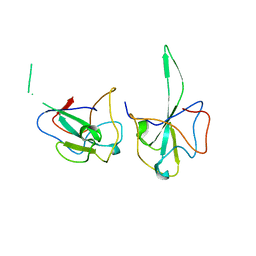 | |
6KL6
 
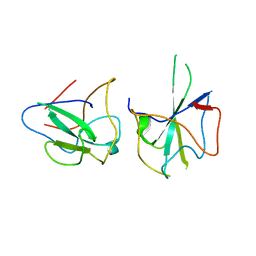 | | Crystal structure of MERS-CoV N-NTD complexed with 5-Benzyloxygramine | | Descriptor: | N,N-dimethyl-1-(5-phenylmethoxy-1H-indol-3-yl)methanamine, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Wang, Y.S, Hsu, J.N. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6KL2
 
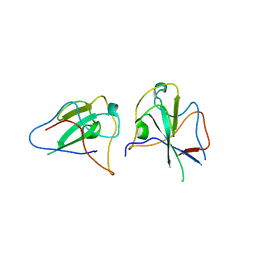 | |
4KC3
 
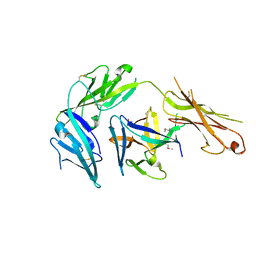 | | Cytokine/receptor binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor-like 1, Interleukin-33 | | Authors: | Liu, X, Wang, X.Q. | | Deposit date: | 2013-04-24 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2702 Å) | | Cite: | Structural insights into the interaction of IL-33 with its receptors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6LNN
 
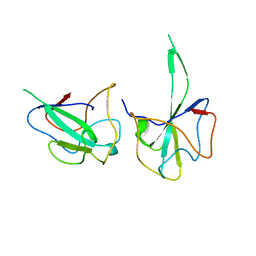 | |
6LZ6
 
 | |
6LZ8
 
 | |
8HHG
 
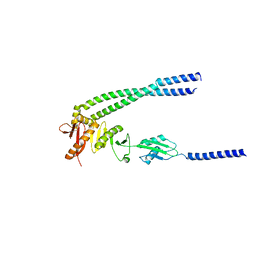 | | The bacterial divisome protein complex FtsB-FtsL-FtsQ | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the heterotrimeric membrane protein complex FtsB-FtsL-FtsQ of the bacterial divisome.
Nat Commun, 14, 2023
|
|
8HHF
 
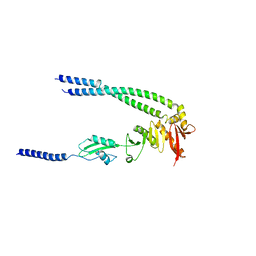 | | The bacterial divisome protein complex FtsB-FtsL-FtsQ | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of the heterotrimeric membrane protein complex FtsB-FtsL-FtsQ of the bacterial divisome.
Nat Commun, 14, 2023
|
|
8HHH
 
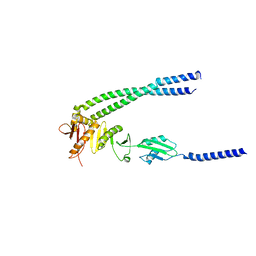 | | The bacterial divisome protein complex FtsB-FtsL-FtsQ | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the heterotrimeric membrane protein complex FtsB-FtsL-FtsQ of the bacterial divisome.
Nat Commun, 14, 2023
|
|
8HYN
 
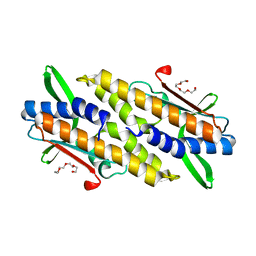 | | Bacterial STING from Riemerella anatipestifer | | Descriptor: | CD-NTase-associated protein 12, TETRAETHYLENE GLYCOL | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-07 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HWJ
 
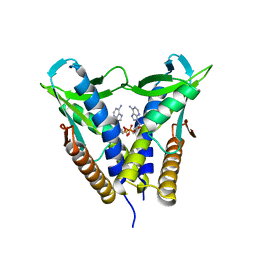 | | Bacterial STING from Epilithonimonas lactis in complex with 3'3'-c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HWI
 
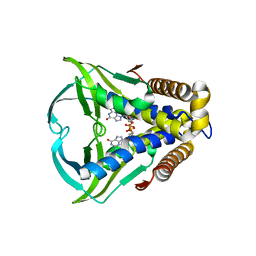 | | Bacterial STING from Larkinella arboricola in complex with 3'3'-c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HY8
 
 | | Bacterial STING from Epilithonimonas lactis | | Descriptor: | CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HY9
 
 | | Bacterial STING from Riemerella anatipestifer in complex with 3'3'-c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.462 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8IBH
 
 | | Cep57 C-terminal domain | | Descriptor: | Centrosomal protein of 57 kDa | | Authors: | Chen, T, Yeh, H.-W, Cheng, H.-C. | | Deposit date: | 2023-02-10 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cep57 regulates human centrosomes through multivalent interactions.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
