4BVK
 
 | |
4BVL
 
 | |
4BVJ
 
 | | Structure of Y105A mutant of PhaZ7 PHB depolymerase | | Descriptor: | PHB DEPOLYMERASE PHAZ7, SODIUM ION | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-06-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
4BRS
 
 | | Structure of wild type PhaZ7 PHB depolymerase | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHB DEPOLYMERASE PHAZ7, ... | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-06-05 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
4BYM
 
 | | Structure of PhaZ7 PHB depolymerase Y105E mutant | | Descriptor: | CHLORIDE ION, PHB DEPOLYMERASE PHAZ7, SODIUM ION | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-07-20 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
4BTV
 
 | | Structure of PhaZ7 PHB depolymerase in complex with 3HB trimer | | Descriptor: | (1R)-3-{[(1R)-3-METHOXY-1-METHYL-3-OXOPROPYL]OXY}-1-METHYL-3-OXOPROPYL (3R)-3-HYDROXYBUTANOATE, PHB DEPOLYMERASE PHAZ7 | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-06-19 | | Release date: | 2013-09-18 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
5MLX
 
 | | Open loop conformation of PhaZ7 Y105E mutant | | Descriptor: | CHLORIDE ION, PHB depolymerase PhaZ7, SODIUM ION | | Authors: | Kellici, T, Mavromoustakos, T, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure analysis, covalent docking, and molecular dynamics calculations reveal a conformational switch in PhaZ7 PHB depolymerase.
Proteins, 85, 2017
|
|
5MLY
 
 | | Closed loop conformation of PhaZ7 Y105E mutant | | Descriptor: | PHB depolymerase PhaZ7 | | Authors: | Kellici, T, Mavromoustakos, T, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure analysis, covalent docking, and molecular dynamics calculations reveal a conformational switch in PhaZ7 PHB depolymerase.
Proteins, 85, 2017
|
|
2VTV
 
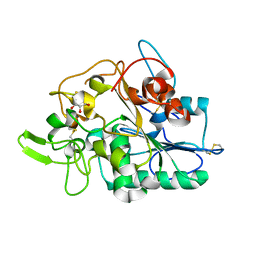 | | PhaZ7 depolymerase from Paucimonas lemoignei | | Descriptor: | GLYCEROL, PHB depolymerase PhaZ7 | | Authors: | Papageorgiou, A.C, Hermawan, S, Singh, C.B, Jendrossek, D. | | Deposit date: | 2008-05-16 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of poly(3-hydroxybutyrate) hydrolysis by PhaZ7 depolymerase from Paucimonas lemoignei.
J. Mol. Biol., 382, 2008
|
|
4ZU2
 
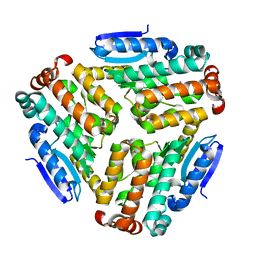 | | Pseudomonas aeruginosa AtuE | | Descriptor: | IODIDE ION, Putative isohexenylglutaconyl-CoA hydratase | | Authors: | Poudel, N, Pfannstiel, J, Simon, O, Walter, N, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Pseudomonas aeruginosa Isohexenyl Glutaconyl Coenzyme A Hydratase (AtuE) Is Upregulated in Citronellate-Grown Cells and Belongs to the Crotonase Family.
Appl.Environ.Microbiol., 81, 2015
|
|
4B2N
 
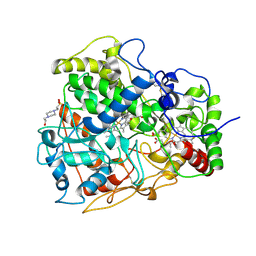 | | Latex Oxygenase RoxA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 70 KDA PROTEIN, HEME C, ... | | Authors: | Seidel, J, Schmitt, G, Hoffmann, M, Jendrossek, D, Einsle, O. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the processive rubber oxygenase RoxA from Xanthomonas sp.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
5O1L
 
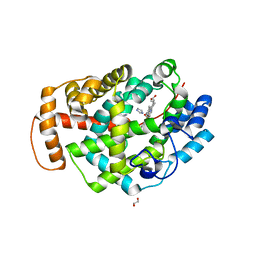 | | Structure of Latex Clearing Protein LCP in the open state with bound imidazole | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, IMIDAZOLE, ... | | Authors: | Ilcu, L, Roether, W, Birke, J, Brausemann, A, Einsle, O, Jendrossek, D. | | Deposit date: | 2017-05-18 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Functional Analysis of Latex Clearing Protein (Lcp) Provides Insight into the Enzymatic Cleavage of Rubber.
Sci Rep, 7, 2017
|
|
5O1M
 
 | | Structure of Latex Clearing Protein LCP in the closed state | | Descriptor: | 1,2-ETHANEDIOL, PROTOPORPHYRIN IX CONTAINING FE, Rubber oxygenase | | Authors: | Ilcu, L, Roether, W, Birke, J, Brausemann, A, Einsle, O, Jendrossek, D. | | Deposit date: | 2017-05-18 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of Latex Clearing Protein (Lcp) Provides Insight into the Enzymatic Cleavage of Rubber.
Sci Rep, 7, 2017
|
|
2X76
 
 | | The crystal structure of PhaZ7 at atomic (1.2 Angstrom) resolution reveals details of the active site and suggests a substrate binding mode | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Wakadkar, S, Hermawan, S, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2010-02-24 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of PhaZ7 at atomic (1.2 A) resolution reveals details of the active site and suggests a substrate-binding mode.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 66, 2010
|
|
2X5X
 
 | | The crystal structure of PhaZ7 at atomic (1.2 Angstrom) resolution reveals details of the active site and suggests a substrate binding mode | | Descriptor: | CHLORIDE ION, IODIDE ION, PHB DEPOLYMERASE PHAZ7, ... | | Authors: | Wakadkar, S, Hermawan, S, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of PhaZ7 at atomic (1.2 A) resolution reveals details of the active site and suggests a substrate-binding mode.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 66, 2010
|
|
