3PIH
 
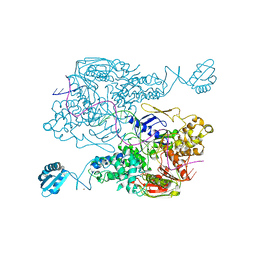 | | T. maritima UvrA in complex with fluorescein-modified DNA | | Descriptor: | DNA (32-MER), PYROPHOSPHATE, UvrABC system protein A, ... | | Authors: | Jaciuk, M, Nowak, E, Nowotny, M. | | Deposit date: | 2010-11-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of UvrA nucleotide excision repair protein in complex with modified DNA.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8AT6
 
 | | Cryo-EM structure of yeast Elp456 subcomplex | | Descriptor: | Elongator complex protein 4, Elongator complex protein 5, Elongator complex protein 6 | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8ASW
 
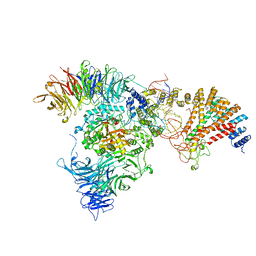 | | Cryo-EM structure of yeast Elp123 in complex with alanine tRNA | | Descriptor: | 5'-DEOXYADENOSINE, Alanine tRNA, Elongator complex protein 1, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-21 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8AVG
 
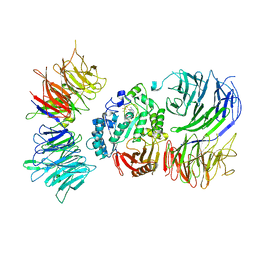 | | Cryo-EM structure of mouse Elp123 with bound SAM | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8ASV
 
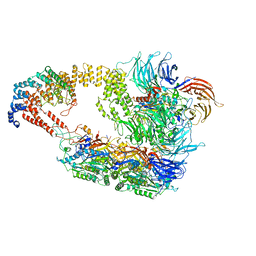 | | Cryo-EM structure of yeast Elongator complex | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-21 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
6QK7
 
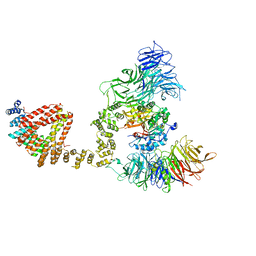 | | Elongator catalytic subcomplex Elp123 lobe | | Descriptor: | 5'-DEOXYADENOSINE, Elongator complex protein 1, Elongator complex protein 2, ... | | Authors: | Dauden, M.I, Jaciuk, M, Glatt, S. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of tRNA recognition by the Elongator complex.
Sci Adv, 5, 2019
|
|
4Q0W
 
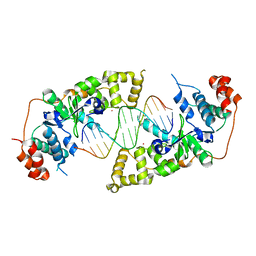 | | he catalytic core of Rad2 in complex with DNA substrate (complex II) | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*TP*AP*GP*GP*TP*GP*GP*AP*CP*GP*GP*AP*TP*CP*AP*TP*T)-3'), DNA (5'-D(*TP*TP*TP*GP*AP*TP*CP*CP*GP*TP*CP*CP*AP*CP*CP*TP*TP*T)-3'), ... | | Authors: | Mietus, M, Nowak, E, Jaciuk, M, Kustosz, P, Nowotny, M. | | Deposit date: | 2014-04-02 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic core of Rad2: insights into the mechanism of substrate binding.
Nucleic Acids Res., 42, 2014
|
|
7QRM
 
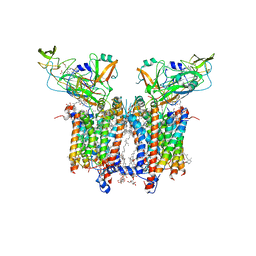 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.7 A resolution | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Sarewicz, M, Szwalec, M, Indyka, P, Rawski, M, Pintscher, S, Pietras, R, Mielecki, B, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-01-11 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
9ES8
 
 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f with decylplastoquinone bound at plastoquionol reduction site | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
9ES9
 
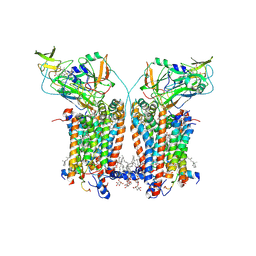 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f complex with inhibitor DBMIB bound at plastoquinol oxidation site | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,5-DIBROMO-3-ISOPROPYL-6-METHYLBENZO-1,4-QUINONE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
9ES7
 
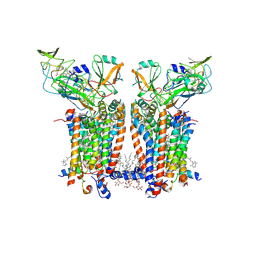 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f complex with water molecules at 1.94 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
4Q0R
 
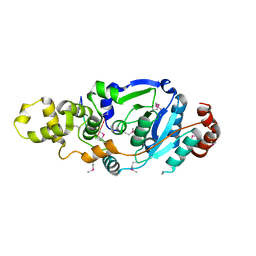 | | The catalytic core of Rad2 (complex I) | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*GP*TP*CP*AP*GP*AP*GP*CP*AP*AP*A)-3'), DNA repair protein RAD2 | | Authors: | Mietus, M, Nowak, E, Jaciuk, M, Kustosz, P, Nowotny, M. | | Deposit date: | 2014-04-02 | | Release date: | 2014-08-27 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the catalytic core of Rad2: insights into the mechanism of substrate binding.
Nucleic Acids Res., 42, 2014
|
|
4Q0Z
 
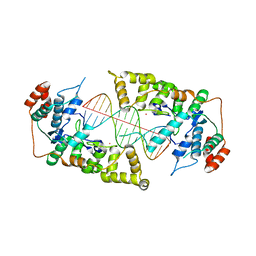 | | The catalytic core of Rad2 in complex with DNA substrate (complex III) | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*CP*TP*GP*AP*GP*AP*CP*AP*AP*GP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*CP*CP*TP*TP*GP*TP*CP*TP*CP*AP*GP*T)-3'), ... | | Authors: | Mietus, M, Nowak, E, Jaciuk, M, Kustosz, P, Nowotny, M. | | Deposit date: | 2014-04-02 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Crystal structure of the catalytic core of Rad2: insights into the mechanism of substrate binding.
Nucleic Acids Res., 42, 2014
|
|
4Q10
 
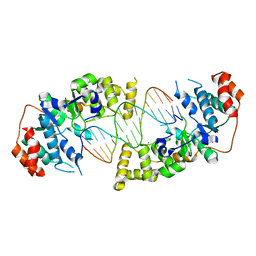 | | The catalytic core of Rad2 in complex with DNA substrate (complex IV) | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*TP*TP*TP*CP*TP*GP*AP*GP*AP*CP*AP*AP*GP*GP*GP*AP*GP*CP*TP*TP*TP*T)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*TP*CP*CP*CP*TP*TP*GP*TP*CP*TP*CP*AP*GP*TP*TP*TP*T)-3'), ... | | Authors: | Mietus, M, Nowak, E, Jaciuk, M, Kustosz, P, Nowotny, M. | | Deposit date: | 2014-04-02 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the catalytic core of Rad2: insights into the mechanism of substrate binding.
Nucleic Acids Res., 42, 2014
|
|
8PU0
 
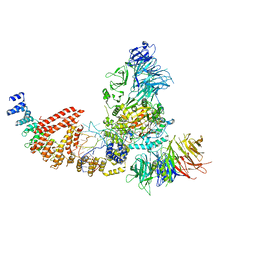 | | Cryo-EM structure of human Elp123 in complex with tRNA, desulpho-CoA, 5'-deoxyadenosine and methionine | | Descriptor: | 5'-DEOXYADENOSINE, DESULFO-COENZYME A, Elongator complex protein 1, ... | | Authors: | Abbassi, N, Jaciuk, M, Lin, T.-Y, Glatt, S. | | Deposit date: | 2023-07-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structures of the human Elongator complex at work.
Nat Commun, 15, 2024
|
|
8PTZ
 
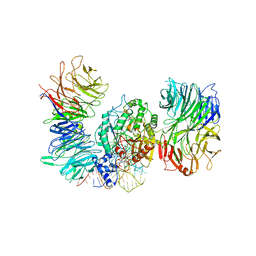 | | Cryo-EM structure of human Elp123 in complex with tRNA, S-ethyl-CoA, 5'-deoxyadenosine and methionine | | Descriptor: | 5'-DEOXYADENOSINE, Elongator complex protein 1, Elongator complex protein 2, ... | | Authors: | Abbassi, N, Jaciuk, M, Lin, T.-Y, Glatt, S. | | Deposit date: | 2023-07-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structures of the human Elongator complex at work.
Nat Commun, 15, 2024
|
|
8PTY
 
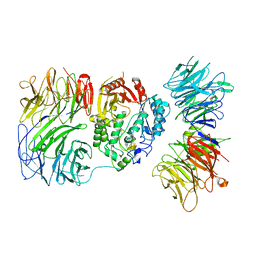 | | Cryo-EM structure of human Elp123 in complex with 5'-deoxyadenosine and methionine | | Descriptor: | 5'-DEOXYADENOSINE, Elongator complex protein 1, Elongator complex protein 2, ... | | Authors: | Abbassi, N, Jaciuk, M, Lin, T.-Y, Glatt, S. | | Deposit date: | 2023-07-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of the human Elongator complex at work.
Nat Commun, 15, 2024
|
|
8PTX
 
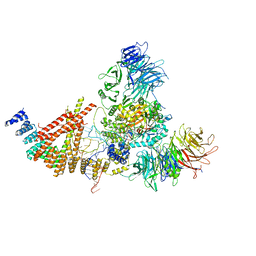 | | Cryo-EM structure of human Elp123 in complex with tRNA, acetyl-CoA, 5'-deoxyadenosine and methionine | | Descriptor: | 5'-DEOXYADENOSINE, ACETYL COENZYME *A, Elongator complex protein 1, ... | | Authors: | Abbassi, N, Jaciuk, M, Lin, T.-Y, Glatt, S. | | Deposit date: | 2023-07-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structures of the human Elongator complex at work.
Nat Commun, 15, 2024
|
|
7ZYV
 
 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.13 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, ... | | Authors: | Sarewicz, M, Szwalec, M, Pintscher, S, Indyka, P, Rawski, M, Pietras, R, Mielecki, B, Koziej, L, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
