1WMB
 
 | | Crystal structure of NAD dependent D-3-hydroxybutylate dehydrogenase | | Descriptor: | CACODYLATE ION, D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION | | Authors: | Ito, K, Nakajima, Y, Ichihara, E, Ogawa, K, Yoshimoto, T. | | Deposit date: | 2004-07-06 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | d-3-Hydroxybutyrate Dehydrogenase from Pseudomonas fragi: Molecular Cloning of the Enzyme Gene and Crystal Structure of the Enzyme
J.Mol.Biol., 355, 2006
|
|
1X1T
 
 | | Crystal Structure of D-3-Hydroxybutyrate Dehydrogenase from Pseudomonas fragi Complexed with NAD+ | | Descriptor: | CACODYLATE ION, D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Ito, K, Nakajima, Y, Ichihara, E, Ogawa, K, Yoshimoto, T. | | Deposit date: | 2005-04-13 | | Release date: | 2006-01-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | d-3-Hydroxybutyrate Dehydrogenase from Pseudomonas fragi: Molecular Cloning of the Enzyme Gene and Crystal Structure of the Enzyme
J.Mol.Biol., 355, 2006
|
|
1V4B
 
 | |
3AIC
 
 | | Crystal Structure of Glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
3AIB
 
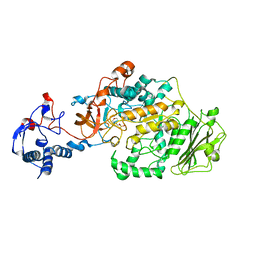 | | Crystal Structure of Glucansucrase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
3AIE
 
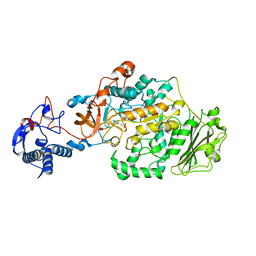 | | Crystal Structure of glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
6M5A
 
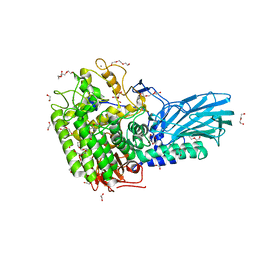 | | Crystal structure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum | | Descriptor: | 1,2-ETHANEDIOL, Beta-L-arabinobiosidase, CALCIUM ION, ... | | Authors: | Saito, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of beta-L-arabinobiosidase belonging to glycoside hydrolase family 121.
Plos One, 15, 2020
|
|
2Z98
 
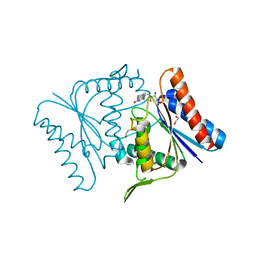 | |
2Z9C
 
 | |
2Z9D
 
 | |
2Z9B
 
 | |
2D5I
 
 | |
2CZC
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Pyrococcus horikoshii OT3 | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Ito, K, Arai, R, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Pyrococcus horikoshii OT3
To be Published
|
|
2CZF
 
 | |
2CYJ
 
 | | Crystal structure of conserved hypothetical protein PH1505 from Pyrococcus horikoshii OT3 | | Descriptor: | ACETATE ION, SULFATE ION, hypothetical protein PH1505 | | Authors: | Ito, K, Arai, R, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-07 | | Release date: | 2006-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of conserved hypothetical protein PH1505 from Pyrococcus horikoshii OT3
To be Published
|
|
2CWQ
 
 | | Crystal structure of conserved protein TTHA0727 from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TTHA0727 | | Authors: | Ito, K, Arai, R, Fusatomi, E, Kamo-Uchikubo, T, Kawaguchi, S, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-23 | | Release date: | 2005-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the conserved protein TTHA0727 from Thermus thermophilus HB8 at 1.9 A resolution: A CMD family member distinct from carboxymuconolactone decarboxylase (CMD) and AhpD
Protein Sci., 15, 2006
|
|
3VJR
 
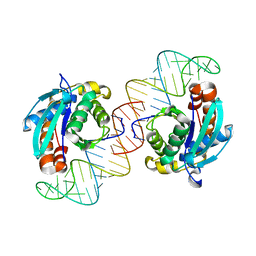 | | Crystal structure of Peptidyl-tRNA hydrolase from Escherichia coli in complex with the CCA-acceptor-T[PSI]C domain of tRNA | | Descriptor: | Peptidyl-tRNA hydrolase, tRNA CCA-acceptor | | Authors: | Ito, K, Murakami, R, Mochizuki, M, Qi, H, Shimizu, Y, Miura, K.I, Ueda, T, Uchiumi, T. | | Deposit date: | 2011-10-28 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the substrate recognition and catalysis of peptidyl-tRNA hydrolase.
Nucleic Acids Res., 40, 2012
|
|
3WY9
 
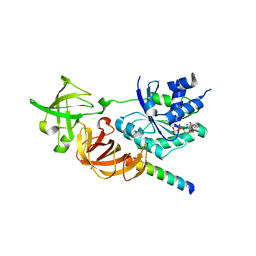 | | Crystal structure of a complex of the archaeal ribosomal stalk protein aP1 and the GDP-bound archaeal elongation factor aEF1alpha | | Descriptor: | 50S ribosomal protein L12, Elongation factor 1-alpha, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ito, K, Honda, T, Suzuki, T, Miyoshi, T, Murakami, R, Yao, M, Uchiumi, T. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1 alpha.
Nucleic Acids Res., 42, 2014
|
|
3WYA
 
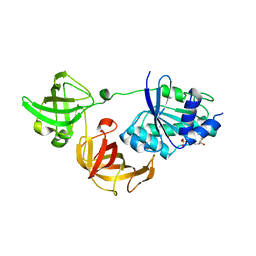 | | Crystal structure of GDP-bound EF1alpha from Pyrococcus horikoshii | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ito, K, Honda, T, Suzuki, T, Miyoshi, T, Murakami, R, Yao, M, Uchiumi, T. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1 alpha.
Nucleic Acids Res., 42, 2014
|
|
1UF0
 
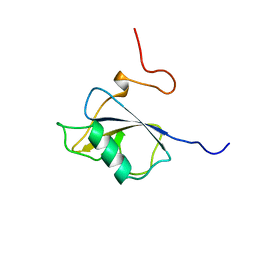 | | Solution structure of the N-terminal DCX domain of human doublecortin-like kinase | | Descriptor: | Serine/threonine-protein kinase DCAMKL1 | | Authors: | Saito, K, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-22 | | Release date: | 2003-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal DCX domain of human doublecortin-like kinase
To be Published
|
|
1V65
 
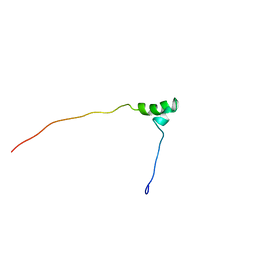 | | Solution structure of the Kruppel-associated box (KRAB) domain | | Descriptor: | RIKEN cDNA 2610044O15 | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Kruppel-associated box (KRAB) domain
To be Published
|
|
1V64
 
 | | Solution structure of the 3rd HMG box of mouse UBF1 | | Descriptor: | Nucleolar transcription factor 1 | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 3rd HMG box of mouse UBF1
To be Published
|
|
1V62
 
 | | Solution structure of the 3rd PDZ domain of GRIP2 | | Descriptor: | KIAA1719 protein | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 3rd PDZ domain of GRIP2
To be Published
|
|
1UF1
 
 | | Solution structure of the second PDZ domain of human KIAA1526 protein | | Descriptor: | KIAA1526 protein | | Authors: | Saito, K, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-22 | | Release date: | 2003-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second PDZ domain of human KIAA1526 protein
To be Published
|
|
1UHS
 
 | | Solution structure of mouse homeodomain-only protein HOP | | Descriptor: | homeodomain only protein | | Authors: | Saito, K, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse homeodomain-only protein HOP
To be Published
|
|
