3X0L
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ES-state at 1.00 angstrom resolution | | Descriptor: | ADP-ribose pyrophosphatase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X0Q
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ESMM-state at reaction time of 20 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X0K
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ES-state at 0.97 angstrom resolution | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-16 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X0I
 
 | | ADP ribose pyrophosphatase in apo state at 0.91 angstrom resolution | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, SULFATE ION | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-16 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X0M
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ESM-state at reaction time of 3 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X0N
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ESM-state at reaction time of 6 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X0P
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ESMM-state at reaction time of 15 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X0R
 
 | | DP ribose pyrophosphatase from Thermus thermophilus HB8 in E'-state at reaction time of 30 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X0O
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ESMM-state at reaction time of 10 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X0J
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in apo state at 0.92 angstrom resolution | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, SULFATE ION | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-16 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X0S
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in E'-state at reaction time of 50 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
8WV8
 
 | |
8WVE
 
 | |
7V3X
 
 | |
7DYK
 
 | |
7DYI
 
 | |
7DYJ
 
 | |
7DXQ
 
 | |
7DYE
 
 | |
7DY2
 
 | |
7DY1
 
 | |
4XZG
 
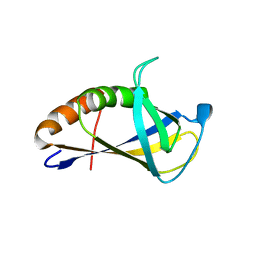 | | Crystal structure of HIRAN domain of human HLTF | | Descriptor: | Helicase-like transcription factor | | Authors: | Ikegaya, Y, Hara, K, Hashimoto, H. | | Deposit date: | 2015-02-04 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Novel DNA-binding Domain of Helicase-like Transcription Factor (HLTF) and Its Functional Implication in DNA Damage Tolerance
J.Biol.Chem., 290, 2015
|
|
7WDC
 
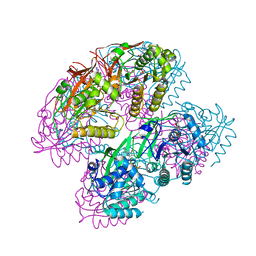 | | Crystal Structure of Cyanobacterial Circadian Clock Protein KaiC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Furuike, Y, Akiyama, S. | | Deposit date: | 2021-12-21 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Highly sensitive tryptophan fluorescence probe for detecting rhythmic conformational changes of KaiC in the cyanobacterial circadian clock system.
Biochem.J., 479, 2022
|
|
7CAO
 
 | | Crystal structure of red chromoprotein from Olindias formosa | | Descriptor: | Chromoprotein | | Authors: | Nakashima, R, Zhai, L, Ike, Y, Matsudz, T, Nagai, T. | | Deposit date: | 2020-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based analysis and evolution of a monomerized red-colored chromoprotein from the Olindias formosa jellyfish.
Protein Sci., 31, 2022
|
|
1UF5
 
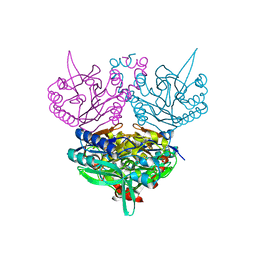 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-methionine | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYLSULFANYL-2-UREIDO-BUTYRIC ACID, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of C171A/V236A mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
