4URE
 
 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, CYCLOHEXANOL DEHYDROGENASE, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
4URF
 
 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, BICARBONATE ION, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
3ZN2
 
 | | protein engineering of halohydrin dehalogenase | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETATE ION, HALOHYDRIN DEHALOGENASE, ... | | Authors: | Schallmey, M, Jekel, P, Tang, L, Majeric-Elenkov, M, Hoeffken, H.W, Hauer, B, Janssen, D.B. | | Deposit date: | 2013-02-13 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Single Point Mutation Enhances Hydroxynitrile Synthesis by Halohydrin Dehalogenase.
Enzyme.Microb.Technol., 70, 2015
|
|
6G71
 
 | | Structure of CYP1232A24 from Arthrobacter sp. | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, FE (III) ION, ... | | Authors: | Dubiel, P, Sharma, M, Klenk, J, Hauer, B, Grogan, G. | | Deposit date: | 2018-04-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and characterization of cytochrome P450 1232A24 and 1232F1 from Arthrobacter sp. and their role in the metabolic pathway of papaverine.
J.Biochem., 166, 2019
|
|
2XEC
 
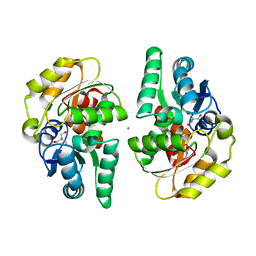 | | Nocardia farcinica maleate cis-trans isomerase bound to TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, PUTATIVE MALEATE ISOMERASE | | Authors: | Fisch, F, Martinez-Fleites, C, Baudendistel, N, Hauer, B, Turkenburg, J.P, Hart, S, Bruce, N.C, Grogan, G. | | Deposit date: | 2010-05-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Covalent Succinylcysteine-Like Intermediate in the Enzyme-Catalyzed Transformation of Maleate to Fumarate by Maleate Isomerase.
J.Am.Chem.Soc., 132, 2010
|
|
2XED
 
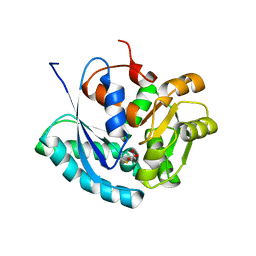 | | Nocardia farcinica maleate cis-trans isomerase C194S mutant with a covalently bound succinylcysteine intermediate | | Descriptor: | PUTATIVE MALEATE ISOMERASE, SUCCINIC ACID | | Authors: | Fisch, F, Martinez-Fleites, C, Baudendistel, N, Hauer, B, Turkenburg, J.P, Hart, S, Bruce, N.C, Grogan, G. | | Deposit date: | 2010-05-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Covalent Succinylcysteine-Like Intermediate in the Enzyme-Catalyzed Transformation of Maleate to Fumarate by Maleate Isomerase.
J.Am.Chem.Soc., 132, 2010
|
|
5FYG
 
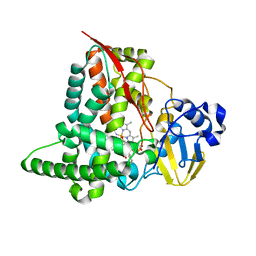 | | Structure of CYP153A from Marinobacter aquaeolei in complex with hydroxydodecanoic acid | | Descriptor: | 12-HYDROXYDODECANOIC ACID, CYTOCHROME P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Danesh-Azari, H.-R, Spandolf, C, Hoffman, S.M, Weissenborn, M, Hauer, B, Grogan, G. | | Deposit date: | 2016-03-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Guided Redesign of CYP153AM.aqfor the Improved Terminal Hydroxylation of Fatty Acids
Chemcatchem, 2016
|
|
5FYF
 
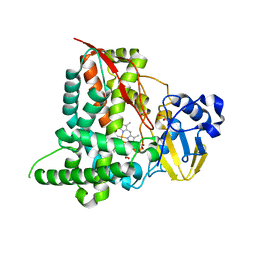 | | Structure of CYP153A from Marinobacter aquaeolei | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Danesh Azari, H.R, Spandolf, C, Hoffman, S.M, Weissenborn, M, Hauer, B, Grogan, G. | | Deposit date: | 2016-03-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-Guided Redesign of CYP153AM.aqfor the Improved Terminal Hydroxylation of Fatty Acids
Chemcatchem, 2016
|
|
4IY1
 
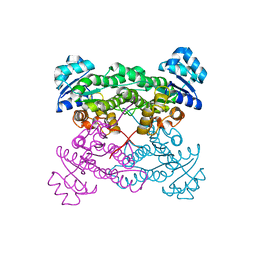 | | Structure of a 37-fold mutant of halohydrin dehalogenase (HheC) with chloride bound | | Descriptor: | CHLORIDE ION, Halohydrin dehalogenase | | Authors: | Floor, R.J, Schallmey, M, Hauer, B, Breuer, M, Jekel, P.A, Wijma, H.J, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biocatalytic and structural properties of a highly engineered halohydrin dehalogenase.
Chembiochem, 14, 2013
|
|
4IXT
 
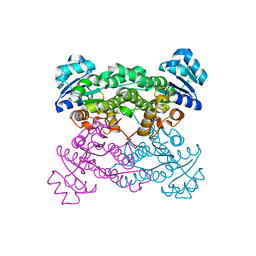 | | Structure of a 37-fold mutant of halohydrin dehalogenase (HheC) bound to ethyl (R)-4-cyano-3-hydroxybutyrate | | Descriptor: | CHLORIDE ION, Halohydrin dehalogenase, ethyl (3R)-4-cyano-3-hydroxybutanoate | | Authors: | Floor, R.J, Schallmey, M, Hauer, B, Breuer, M, Jekel, P.A, Wijma, H.J, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Biocatalytic and structural properties of a highly engineered halohydrin dehalogenase.
Chembiochem, 14, 2013
|
|
4IXW
 
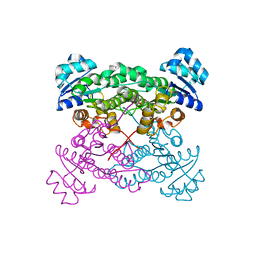 | | Halohydrin dehalogenase (HheC) bound to ethyl (2S)-oxiran-2-ylacetate | | Descriptor: | CHLORIDE ION, Halohydrin dehalogenase, ethyl (2S)-oxiran-2-ylacetate | | Authors: | Floor, R.J, Schallmey, M, Hauer, B, Breuer, M, Jekel, P.A, Wijma, H.J, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Biocatalytic and structural properties of a highly engineered halohydrin dehalogenase.
Chembiochem, 14, 2013
|
|
7ANT
 
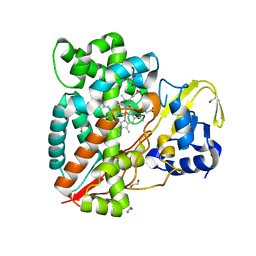 | | Structure of CYP153A from Polaromonas sp. | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, HEME C | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffman, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-12 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
7AO7
 
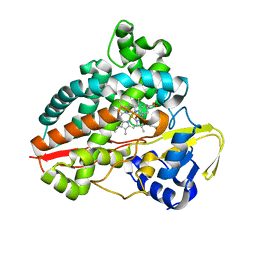 | | Structure of CYP153A from Polaromonas sp. in complex with octan-1-ol | | Descriptor: | Cytochrome P450, OCTAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffmann, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
6GMF
 
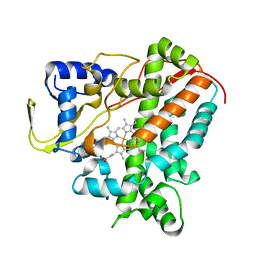 | | Structure of Cytochrome P450 CYP109Q5 from Chondromyces apiculatus | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase | | Authors: | Grogan, G, Dubiel, P, Sharma, M, Klenk, J, Hauer, B. | | Deposit date: | 2018-05-25 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Characterization and structure-guided engineering of the novel versatile terpene monooxygenase CYP109Q5 from Chondromyces apiculatus DSM436.
Microb Biotechnol, 12, 2019
|
|
7AD2
 
 | |
4A3U
 
 | |
3DG9
 
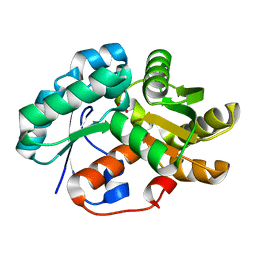 | | Crystal Structure of Malonate Decarboxylase from Bordatella bronchiseptica | | Descriptor: | Arylmalonate decarboxylase, PHOSPHATE ION | | Authors: | Okrasa, K, Levy, C, Baudendistel, N, Leys, D, Micklefield, J. | | Deposit date: | 2008-06-13 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Mechanism of an Unusual Malonate Decarboxylase and Related Racemases.
Chemistry, 14, 2008
|
|
3IP8
 
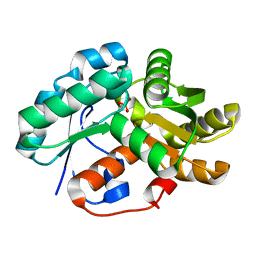 | | Crystal structure of arylmalonate decarboxylase (AMDase) from Bordatella bronchiseptic in complex with benzylphosphonate | | Descriptor: | Arylmalonate decarboxylase, benzylphosphonic acid | | Authors: | Okrasa, K, Levy, C, Leys, D, Micklefield, J. | | Deposit date: | 2009-08-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure-Guided Directed Evolution of Alkenyl and Arylmalonate Decarboxylases.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2EWM
 
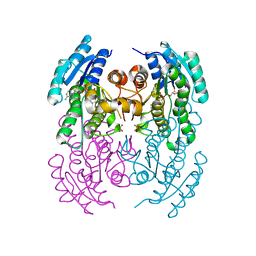 | |
2EW8
 
 | |
4OT7
 
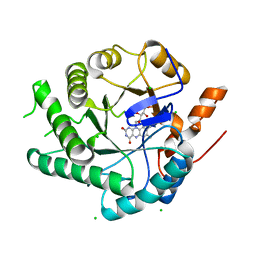 | | X-structure of a variant of NCR from zymomonas mobilis | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase/NADH oxidase | | Authors: | Genz, M, Strater, N. | | Deposit date: | 2014-02-13 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Surface Loops Representing Enzyme Modifying Element in the Field of Tuning Enzyme Properties
TO BE PUBLISHED
|
|
5G1W
 
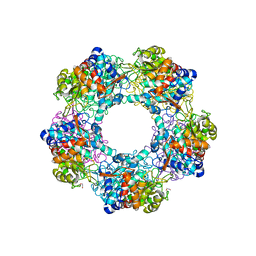 | | Apo Structure of Linalool Dehydratase-Isomerase | | Descriptor: | 1,2-ETHANEDIOL, LINALOOL DEHYDRATASE/ISOMERASE, METHYLMALONIC ACID | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5G1U
 
 | | Linalool Dehydratase Isomerase in complex with Geraniol | | Descriptor: | Geraniol, LINALOOL DEHYDRATASE/ISOMERASE | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5G1V
 
 | | Linalool Dehydratase Isomerase: Selenomethionine Derivative | | Descriptor: | LINALOOL DEHYDRATASE ISOMERASE | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
