2L3Z
 
 | | Proton-Detected 4D DREAM Solid-State NMR Structure of Ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Huber, M, Hiller, S, Schanda, P, Ernst, M, Bockmann, A, Verel, R, Meier, B.H. | | Deposit date: | 2010-09-27 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | A Proton-Detected 4D Solid-State NMR Experiment for Protein Structure Determination.
Chemphyschem, 12, 2011
|
|
2LBU
 
 | | HADDOCK calculated model of Congo red bound to the HET-s amyloid | | Descriptor: | Small s protein, sodium 3,3'-(1E,1'E)-biphenyl-4,4'-diylbis(diazene-2,1-diyl)bis(4-aminonaphthalene-1-sulfonate) | | Authors: | Schutz, A.K, Soragni, A, Hornemann, S, Aguzzi, A, Ernst, M, Bockmann, A, Meier, B.H. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The Amyloid-Congo Red Interface at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 2011
|
|
2XNF
 
 | | The Mediator Med25 activator interaction domain: Structure and cooperative binding of VP16 subdomains | | Descriptor: | MEDIATOR OF RNA POLYMERASE II TRANSCRIPTION SUBUNIT 25 | | Authors: | Vojnic, E, Mourao, A, Seizl, M, Simon, B, Wenzeck, L, Lariviere, L, Baumli, S, Meisterernst, M, Sattler, M, Cramer, P. | | Deposit date: | 2010-08-02 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Vp16 Binding of the Mediator Med25 Activator Interaction Domain.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1JFI
 
 | | Crystal Structure of the NC2-TBP-DNA Ternary Complex | | Descriptor: | 5'-D(*G*GP*AP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*TP*CP*C)-3', TATA-BOX-BINDING PROTEIN (TBP), ... | | Authors: | Kamada, K, Shu, F, Chen, H, Malik, S, Stelzer, G, Roeder, R.G, Meisterernst, M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-06-20 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex.
Cell(Cambridge,Mass.), 106, 2001
|
|
2YOE
 
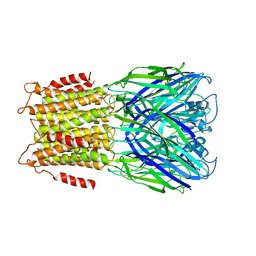 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with GABA and flurazepam | | Descriptor: | CYS-LOOP LIGAND-GATED ION CHANNEL, Flurazepam, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Spurny, R, Brams, M, Nury, H, Legrand, P, Ulens, C. | | Deposit date: | 2012-10-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Pentameric Ligand-Gated Ion Channel Elic is Activated by Gaba and Modulated by Benzodiazepines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4A97
 
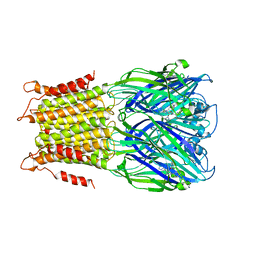 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with zopiclone | | Descriptor: | (5R)-6-(5-chloropyridin-2-yl)-7-oxo-6,7-dihydro-5H-pyrrolo[3,4-b]pyrazin-5-yl 4-methylpiperazine-1-carboxylate, CYS-LOOP LIGAND-GATED ION CHANNEL | | Authors: | Spurny, R, Brams, M, Ulens, C. | | Deposit date: | 2011-11-24 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.343 Å) | | Cite: | Pentameric Ligand-Gated Ion Channel Elic is Activated by Gaba and Modulated by Benzodiazepines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4A98
 
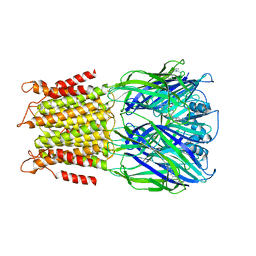 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromoflurazepam | | Descriptor: | 7-BROMO-1-[2-(DIETHYLAMINO)ETHYL]-5-(2-FLUOROPHENYL)-1,3-DIHYDRO-2H-1,4-BENZODIAZEPIN-2-ONE, CYS-LOOP LIGAND-GATED ION CHANNEL | | Authors: | Spurny, R, Brams, M, Nury, H, Legrand, P, Ulens, C. | | Deposit date: | 2011-11-24 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Pentameric Ligand-Gated Ion Channel Elic is Activated by Gaba and Modulated by Benzodiazepines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5JZN
 
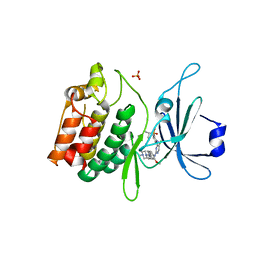 | | Crystal structure of DCLK1-KD in complex with NVP-TAE684 | | Descriptor: | 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE, SULFATE ION, Serine/threonine-protein kinase DCLK1 | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Biochemical and Structural Insights into Doublecortin-like Kinase Domain 1.
Structure, 24, 2016
|
|
5JZJ
 
 | | Crystal structure of DCLK1-KD in complex with AMPPN | | Descriptor: | AMP PHOSPHORAMIDATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Biochemical and Structural Insights into Doublecortin-like Kinase Domain 1.
Structure, 24, 2016
|
|
5L0Q
 
 | | Crystal structure of the complex between ADAM10 D+C domain and a conformation specific mAb 8C7. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Disintegrin and metalloproteinase domain-containing protein 10, MAGNESIUM ION, ... | | Authors: | Xu, K, Saha, N, Nikolov, D.B. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | An activated form of ADAM10 is tumor selective and regulates cancer stem-like cells and tumor growth.
J.Exp.Med., 213, 2016
|
|
2MUS
 
 | | HADDOCK calculated model of LIN5001 bound to the HET-s amyloid | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Heterokaryon incompatibility protein s | | Authors: | Hermann, U.S, Schuetz, A.K, Shirani, H, Saban, D, Nuvolone, M, Huang, D.H, Li, B, Ballmer, B, Aslund, A.K.O, Mason, J.J, Rushing, E, Budka, H, Hammarstrom, P, Bockmann, A, Caflisch, A, Meier, B.H, Nilsson, P.K.R, Hornemann, S, Aguzzi, A. | | Deposit date: | 2014-09-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design identifies polythiophenes as antiprion compounds.
Sci Transl Med, 7, 2015
|
|
1EIJ
 
 | | NMR ENSEMBLE OF METHANOBACTERIUM THERMOAUTOTROPHICUM PROTEIN 1615 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1615 | | Authors: | Christendat, D, Booth, V, Gernstein, M, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-25 | | Release date: | 2000-11-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
1PCF
 
 | |
