5HEW
 
 | | Pentameric ligand-gated ion channel ELIC mutant T28D | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Engeler, S, Dutzler, R. | | Deposit date: | 2016-01-06 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Signal Transduction at the Domain Interface of Prokaryotic Pentameric Ligand-Gated Ion Channels.
Plos Biol., 14, 2016
|
|
7QXF
 
 | |
5HEJ
 
 | |
5HEO
 
 | |
5HEU
 
 | |
5HEH
 
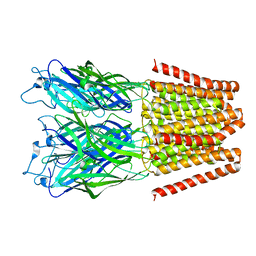 | |
5HEG
 
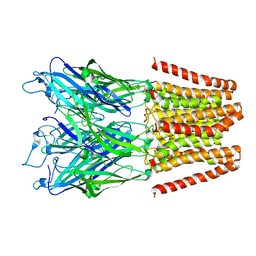 | |
4J8Y
 
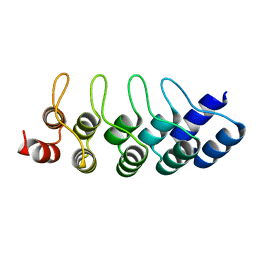 | | E3_5 DARPin D77S mutant | | Descriptor: | DARPin_E3_5_D77S | | Authors: | Seeger, M.A, Gruetter, M.G. | | Deposit date: | 2013-02-15 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity.
Protein Sci., 22, 2013
|
|
4J7W
 
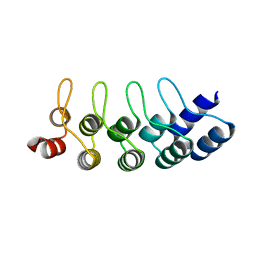 | | E3_5 DARPin L86A mutant | | Descriptor: | DARPin_E3_5_L86A | | Authors: | Seeger, M.A, Gruetter, M.G. | | Deposit date: | 2013-02-14 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity.
Protein Sci., 22, 2013
|
|
4JB8
 
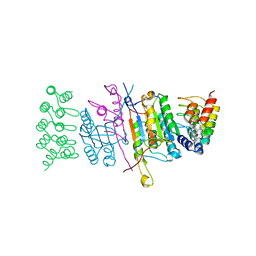 | | Caspase-7 in Complex with DARPin C7_16 | | Descriptor: | Caspase-7 subunit p11, Caspase-7 subunit p20, DARPin C7_16 | | Authors: | Fluetsch, A, Seeger, M.A, Gruetter, M.G. | | Deposit date: | 2013-02-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity.
Protein Sci., 22, 2013
|
|
1YX3
 
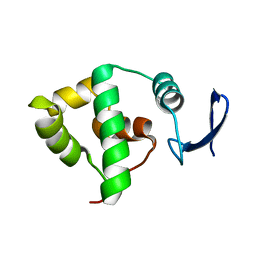 | | NMR structure of Allochromatium vinosum DsrC: Northeast Structural Genomics Consortium target OP4 | | Descriptor: | hypothetical protein DsrC | | Authors: | Cort, J.R, Dahl, C, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-19 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Allochromatium vinosum DsrC: solution-state NMR structure, redox properties, and interaction with DsrEFH, a protein essential for purple sulfur bacterial sulfur oxidation.
J.Mol.Biol., 382, 2008
|
|
