5W1O
 
 | | Crystal Structure of HPV16 L1 Pentamer Bound to Heparin Oligosaccharides | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, Major capsid protein L1 | | Authors: | Dasgupta, J, Chen, X.S. | | Deposit date: | 2017-06-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of oligosaccharide receptor recognition by human papillomavirus.
J. Biol. Chem., 286, 2011
|
|
6LUF
 
 | |
6LUA
 
 | |
1NQP
 
 | | Crystal structure of Human hemoglobin E at 1.73 A resolution | | Descriptor: | CYANIDE ION, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Dasgupta, J, Sen, U, Choudhury, D, Dutta, P, Basu, S, Chakrabarti, A, Chakrabarty, A, Dattagupta, J.K. | | Deposit date: | 2003-01-22 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystallization and preliminary X-ray structural Studies of Hemoglobin A2 and Hemoglobin E, isolated from the blood samples of Beta-thalassemic patients
Biochem.Biophys.Res.Commun., 303, 2004
|
|
4LX8
 
 | |
2ET2
 
 | | Crystal structure of an Asn to Ala mutant of Winged Bean Chymotrypsin Inhibitor protein | | Descriptor: | Chymotrypsin inhibitor 3, NICKEL (II) ION, SULFATE ION | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
2ESU
 
 | | Crystal structure of Asn to Gln mutant of Winged Bean Chymotrypsin Inhibitor protein | | Descriptor: | Chymotrypsin inhibitor 3, NICKEL (II) ION, SULFATE ION | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
2BEA
 
 | | Crystal structure of Asn14 to Gly mutant of WCI | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-24 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
2BEB
 
 | | X-ray structure of Asn to Thr mutant of Winged Bean Chymotrypsin inhibitor | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-24 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
8X1A
 
 | | Crystal structure of periplasmic G6P binding protein VcA0625 | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, Iron(III) ABC transporter, periplasmic iron-compound-binding protein | | Authors: | Dasgupta, J, Saha, I. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural insights in to the atypical type-I ABC Glucose-6-phosphate importer VCA0625-27 of Vibrio cholerae.
Biochem.Biophys.Res.Commun., 716, 2024
|
|
8H5V
 
 | | Crystal structure of the FleQ domain of Vibrio cholerae FlrA | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, flagellar regulatory protein A | | Authors: | Dasgupta, J, Chakraborty, S. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The N-terminal FleQ domain of the Vibrio cholerae flagellar master regulator FlrA plays pivotal structural roles in stabilizing its active state.
Febs Lett., 597, 2023
|
|
7YRT
 
 | |
3TO5
 
 | |
5W1X
 
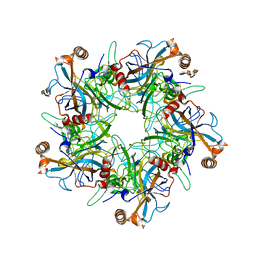 | | Crystal Structure of Humanpapillomavirus18 (HPV18) Capsid L1 Pentamers Bound to Heparin Oligosaccharides | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, ... | | Authors: | Chen, X.S, Dasgupta, J. | | Deposit date: | 2017-06-05 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.374 Å) | | Cite: | Structural basis of oligosaccharide receptor recognition by human papillomavirus.
J. Biol. Chem., 286, 2011
|
|
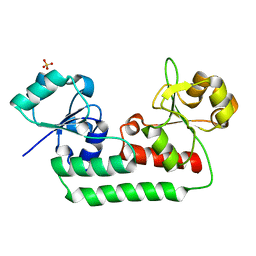
5KHL
 
 | |
1XG6
 
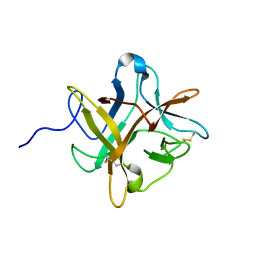 | |
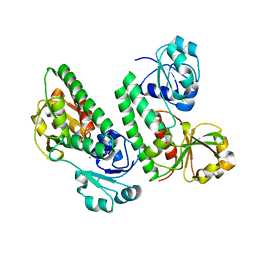
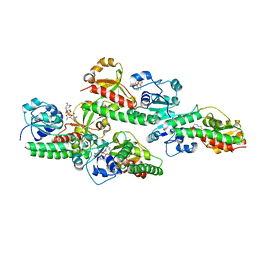
5GGY
 
 | |
5GGX
 
 | |
1SHR
 
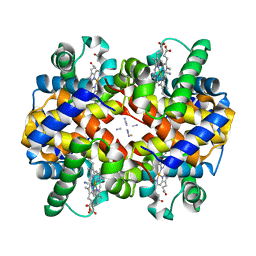 | | Crystal structure of ferrocyanide bound human hemoglobin A2 at 1.88A resolution | | Descriptor: | CYANIDE ION, FE (III) ION, Hemoglobin alpha chain, ... | | Authors: | Sen, U, Dasgupta, J, Choudhury, D, Datta, P, Chakrabarti, A, Chakrabarty, S.B, Chakrabarty, A, Dattagupta, J.K. | | Deposit date: | 2004-02-26 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of HbA2 and HbE and modeling of hemoglobin delta4: interpretation of the thermal stability and the antisickling effect of HbA2 and identification of the ferrocyanide binding site in Hb
Biochemistry, 43, 2004
|
|
1SI4
 
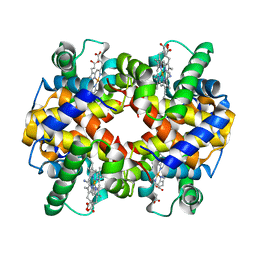 | | Crystal structure of Human hemoglobin A2 (in R2 state) at 2.2 A resolution | | Descriptor: | CYANIDE ION, Hemoglobin alpha chain, Hemoglobin delta chain, ... | | Authors: | Sen, U, Dasgupta, J, Choudhury, D, Datta, P, Chakrabarti, A, Chakrabarty, S.B, Chakrabarty, A, Dattagupta, J.K. | | Deposit date: | 2004-02-27 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of HbA2 and HbE and modeling of hemoglobin delta4: interpretation of the thermal stability and the antisickling effect of HbA2 and identification of the ferrocyanide binding site in Hb.
Biochemistry, 43, 2004
|
|
1FN0
 
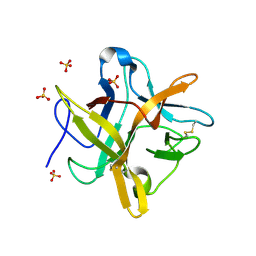 | | STRUCTURE OF A MUTANT WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN, N14D. | | Descriptor: | CHYMOTRYPSIN INHIBITOR 3, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Dasgupta, J, Ghosh, S. | | Deposit date: | 2000-08-19 | | Release date: | 2001-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
PROTEIN ENG., 14, 2001
|
|
3QYD
 
 | | Crystal structure of a recombinant chimeric trypsin inhibitor | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Sen, U, Majumder, S, Khamrui, S, Dasgupta, J. | | Deposit date: | 2011-03-03 | | Release date: | 2011-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Role of remote scaffolding residues in the inhibitory loop pre-organization, flexibility, rigidification and enzyme inhibition of serine protease inhibitors
Biochim.Biophys.Acta, 1824, 2012
|
|
3VEQ
 
 | | A binary complex betwwen bovine pancreatic trypsin and a engineered mutant trypsin inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, Chymotrypsin inhibitor 3 | | Authors: | Sen, U, Majumder, S, Khamrui, S, Dasgupta, J. | | Deposit date: | 2012-01-09 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of remote scaffolding residues in the inhibitory loop pre-organization, flexibility, rigidification and enzyme inhibition of serine protease inhibitors
Biochim.Biophys.Acta, 1824, 2012
|
|
4H60
 
 | |
4QHS
 
 | | Crystal structure of AAA+sigma 54 activator domain of the flagellar regulatory protein FlrC of Vibrio cholerae in nucleotide free state | | Descriptor: | 1,2-ETHANEDIOL, Flagellar regulatory protein C | | Authors: | Dey, S, Biswas, M, Sen, U, Dasgupta, J. | | Deposit date: | 2014-05-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique ATPase site architecture triggers cis-mediated synchronized ATP binding in heptameric AAA+-ATPase domain of flagellar regulatory protein FlrC
J.Biol.Chem., 290, 2015
|
|
