8I8R
 
 | | Cryo-EM Structure of OmpC3-MlaA Complex in MSP2N2 Nanodiscs | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Intermembrane phospholipid transport system lipoprotein MlaA, Outer membrane porin C | | Authors: | Yeow, J, Luo, M, Chng, S.S. | | Deposit date: | 2023-02-05 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Molecular mechanism of phospholipid transport at the bacterial outer membrane interface.
Nat Commun, 14, 2023
|
|
8I8X
 
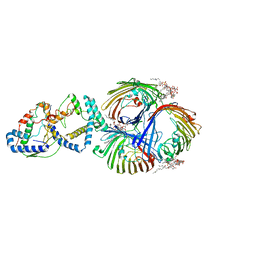 | | Cryo-EM Structure of OmpC3-MlaA-MlaC Complex in MSP2N2 Nanodiscs | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Intermembrane phospholipid transport system binding protein MlaC, Intermembrane phospholipid transport system lipoprotein MlaA, ... | | Authors: | Yeow, J, Luo, M, Chng, S.S. | | Deposit date: | 2023-02-05 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular mechanism of phospholipid transport at the bacterial outer membrane interface.
Nat Commun, 14, 2023
|
|
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
6SAR
 
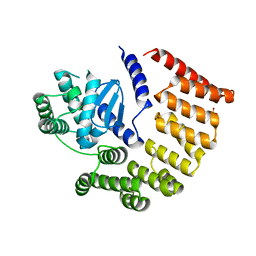 | | E coli BepA/YfgC | | Descriptor: | Beta-barrel assembly-enhancing protease, SULFATE ION, ZINC ION | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2019-07-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Function Characterization of the Conserved Regulatory Mechanism of the Escherichia coli M48 Metalloprotease BepA.
J.Bacteriol., 203, 2020
|
|
