3KFU
 
 | | Crystal structure of the transamidosome | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Glutamyl-tRNA(Gln) amidotransferase subunit A, Glutamyl-tRNA(Gln) amidotransferase subunit C, ... | | Authors: | Blaise, M, Bailly, M, Frechin, M, Thirup, S, Becker, H.D, Kern, D. | | Deposit date: | 2009-10-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation.
Embo J., 29, 2010
|
|
5T2V
 
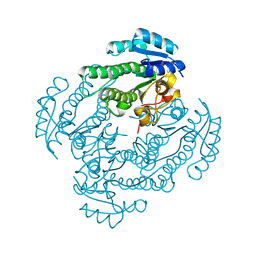 | | Crystal structure of MSMEG_6753 a putative betaketoacyl-ACP reductase | | Descriptor: | MAGNESIUM ION, Oxidoreductase, short chain dehydrogenase/reductase family protein | | Authors: | Blaise, M, Van Wyk, N, Baneres-Roquet, F, Guerardel, Y, Kremer, L. | | Deposit date: | 2016-08-24 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of NADP(+) triggers an open-to-closed transition in a mycobacterial FabG beta-ketoacyl-ACP reductase.
Biochem. J., 474, 2017
|
|
3P8Y
 
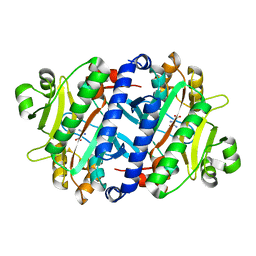 | |
3P8T
 
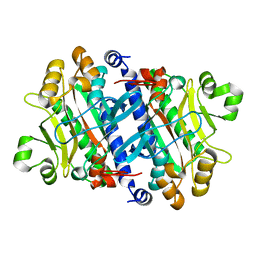 | | Crystal structure of the archaeal asparagine synthetase A | | Descriptor: | AsnS-like asparaginyl-tRNA synthetase related protein | | Authors: | Blaise, M, Kern, D. | | Deposit date: | 2010-10-15 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Archaeal Asparagine Synthetase: Interrelation with Aspartyl-tRNA and Asparaginyl-tRNA Synthetases.
J.Mol.Biol., 412, 2011
|
|
3P8V
 
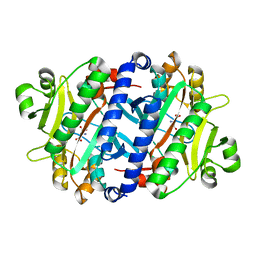 | |
3REX
 
 | | Crystal structure of the archaeal asparagine synthetase A complexed with Adenosine monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, AsnS-like asparaginyl-tRNA synthetase related protein, MAGNESIUM ION | | Authors: | Blaise, M, Frechin, M, Charron, C, Roy, H, Sauter, C, Lorber, B, Olieric, V, Kern, D. | | Deposit date: | 2011-04-05 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Archaeal Asparagine Synthetase: Interrelation with Aspartyl-tRNA and Asparaginyl-tRNA Synthetases.
J.Mol.Biol., 412, 2011
|
|
3RL6
 
 | | Crystal structure of the archaeal asparagine synthetase A complexed with L-Asparagine and Adenosine monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARAGINE, Archaeal asparagine synthetase A, ... | | Authors: | Blaise, M, Frechin, M, Charron, C, Roy, H, Sauter, C, Lorber, B, Olieric, V, Kern, D. | | Deposit date: | 2011-04-19 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Archaeal Asparagine Synthetase: Interrelation with Aspartyl-tRNA and Asparaginyl-tRNA Synthetases.
J.Mol.Biol., 412, 2011
|
|
3REU
 
 | | Crystal structure of the archaeal asparagine synthetase A complexed with L-Aspartic acid and Adenosine triphosphate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTIC ACID, AsnS-like asparaginyl-tRNA synthetase related protein, ... | | Authors: | Blaise, M, Frechin, M, Charron, C, Roy, H, Sauter, C, Lorber, B, Olieric, V, Kern, D. | | Deposit date: | 2011-04-05 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Archaeal Asparagine Synthetase: Interrelation with Aspartyl-tRNA and Asparaginyl-tRNA Synthetases.
J.Mol.Biol., 412, 2011
|
|
4A91
 
 | | Crystal structure of the glutamyl-queuosine tRNAAsp synthetase from E. coli complexed with L-glutamate | | Descriptor: | GLUTAMIC ACID, GLUTAMYL-Q TRNA(ASP) SYNTHETASE, ZINC ION | | Authors: | Blaise, M, Olieric, V, Sauter, C, Lorber, B, Roy, B, Karmakar, S, Banerjee, R, Becker, H.D, Kern, D. | | Deposit date: | 2011-11-22 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Glutamyl-Queuosine Trnaasp Synthetase Complexed with L-Glutamate: Structural Elements Mediating tRNA-Independent Activation of Glutamate and Glutamylation of Trnaasp Anticodon.
J.Mol.Biol., 381, 2008
|
|
4A1K
 
 | | Ykud L,D-transpeptidase | | Descriptor: | PUTATIVE L, D-TRANSPEPTIDASE YKUD, SULFATE ION | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of Three New Crystal Forms of the Ykud L,D-Transpeptidase from B. Subtilis.
To be Published
|
|
4ACJ
 
 | | Crystal structure of the TLDC domain of Oxidation resistance protein 2 from zebrafish | | Descriptor: | WU:FB25H12 PROTEIN, | | Authors: | Blaise, M, B Alsarraf, H.M.A, Wong, J.E.M.M, Midtgaard, S.R, Laroche, F, Schack, L, Spaink, H, Stougaard, J, Thirup, S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal Structure of the Tldc Domain of Oxidation Resistance Protein 2 from Zebrafish.
Proteins, 80, 2012
|
|
4A1J
 
 | | Ykud L,D-transpeptidase from B.subtilis | | Descriptor: | PUTATIVE L, D-TRANSPEPTIDASE YKUD, SULFATE ION | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Ykud
To be Published
|
|
4A1I
 
 | | ykud from B.subtilis | | Descriptor: | CADMIUM ION, CHLORIDE ION, PUTATIVE L, ... | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of Three New Crystal Forms of the Ykud L,D-Transpeptidase from B. Subtilis.
To be Published
|
|
4V12
 
 | | Crystal structure of the MSMEG_6754 dehydratase from Mycobacterium smegmatis | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAOC LIKE DOMAIN PROTEIN | | Authors: | Blaise, M. | | Deposit date: | 2014-09-23 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A New Dehydratase Conferring Innate Resistance to Thiacetazone and Intra-Amoebal Survival of Mycobacterium Smegmatis.
Mol.Microbiol., 96, 2015
|
|
7OBP
 
 | |
7Q3A
 
 | | Crystal structure of MAB_4324 a tandem repeat GNAT from Mycobacterium abscessus | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Blaise, M, Alsarraf, M.A.B. | | Deposit date: | 2021-10-27 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical, structural, and functional studies reveal that MAB_4324c from Mycobacterium abscessus is an active tandem repeat N-acetyltransferase.
Febs Lett., 596, 2022
|
|
5I7N
 
 | | MaoC-like dehydratase | | Descriptor: | MaoC-like dehydratase | | Authors: | Blaise, M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deletion of a dehydratase important for intracellular growth and cording renders rough Mycobacterium abscessus avirulent.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6YCA
 
 | | Crystal structure of Eis1 from Mycobacterium abscessus | | Descriptor: | ACETYL COENZYME *A, SULFATE ION, Uncharacterized N-acetyltransferase D2E76_00625 | | Authors: | Blaise, M, Ung, K.L. | | Deposit date: | 2020-03-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of the N-acetyltransferase Eis1 from Mycobacterium abscessus reveals the molecular determinants of its incapacity to modify aminoglycosides.
Proteins, 89, 2021
|
|
6RFT
 
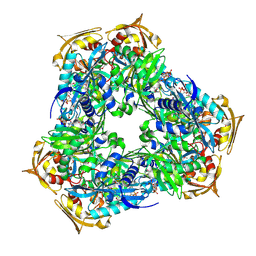 | | Crystal structure of Eis2 from Mycobacterium abscessus bound to Acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Uncharacterized N-acetyltransferase D2E36_21790 | | Authors: | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
6RFX
 
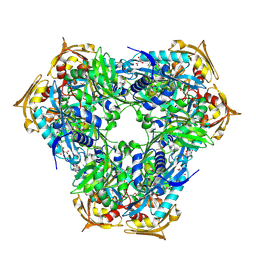 | | Crystal structure of Eis2 from Mycobacterium abscessus | | Descriptor: | ACETATE ION, CITRIC ACID, Eis2, ... | | Authors: | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
6RFY
 
 | | Crystal structure of Eis2 form Mycobacterium abscessus | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Eis2, SULFATE ION | | Authors: | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
6T84
 
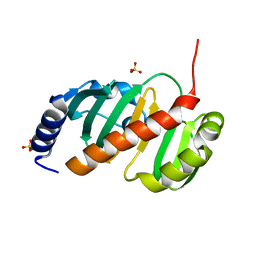 | |
7AGO
 
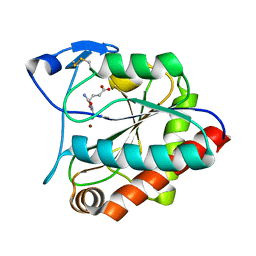 | | crystal structure of the N-acetylmuramyl-L-alanine amidase, Ami1, from Mycobacterium abscessus bound to L-Alanine-D-isoglutamine | | Descriptor: | ALANINE, D-alpha-glutamine, N-acetylmuramoyl-L-alanine amidase, ... | | Authors: | Blaise, M. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Characterization of the N -Acetylmuramyl-l-Alanine Amidase, Ami1, from Mycobacterium abscessus .
Cells, 9, 2020
|
|
7AGM
 
 | | Crystal structure of the N-acetylmuramyl-L-alanine amidase, Ami1, from Mycobacterium smegmatis | | Descriptor: | N-acetylmuramoyl-L-alanine amidase, ZINC ION | | Authors: | Blaise, M, Alsarraf, M.A.B. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Functional Characterization of the N -Acetylmuramyl-l-Alanine Amidase, Ami1, from Mycobacterium abscessus .
Cells, 9, 2020
|
|
7AGL
 
 | |
