3PDX
 
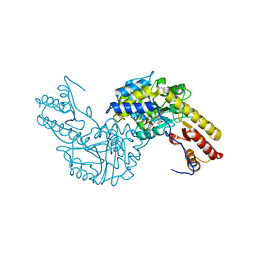 | | Crystal structural of mouse tyrosine aminotransferase | | Descriptor: | Tyrosine aminotransferase | | Authors: | Mehere, P.V, Han, Q, Lemkul, J.A, Robinson, H, Bevan, D.R, Li, J. | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Tyrosine aminotransferase: biochemical and structural properties and molecular dynamics simulations.
Protein Cell, 1, 2010
|
|
3PME
 
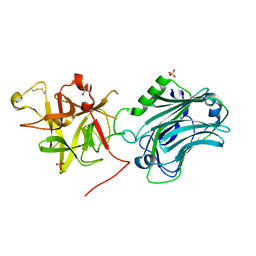 | | Crystal structure of the receptor binding domain of botulinum neurotoxin C/D mosaic serotype | | Descriptor: | GLYCEROL, SULFATE ION, Type C neurotoxin | | Authors: | Zhang, Y, Buchko, G.W, Qin, L, Robinson, H, Varnum, S.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2010-11-16 | | Release date: | 2010-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the receptor binding domain of the botulinum C-D mosaic neurotoxin reveals potential roles of lysines 1118 and 1136 in membrane interactions.
Biochem.Biophys.Res.Commun., 404, 2011
|
|
3PN7
 
 | | Visualizing new hinges and a potential major source of compliance in the lever arm of myosin | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Brown, J.H, Senthil-Kumar, V.S, O'Neall-Hennessey, E, Reshetnikova, L, Robinson, H, Nguyen-McCarty, M, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Visualizing key hinges and a potential major source of compliance in the lever arm of myosin.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2R2N
 
 | | The crystal structure of human kynurenine aminotransferase II in complex with kynurenine | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, GLYCEROL, ... | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2007-08-27 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human kynurenine aminotransferase II.
J.Biol.Chem., 283, 2008
|
|
2R5C
 
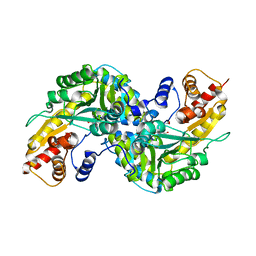 | | Aedes Kynurenine Aminotransferase in Complex with Cysteine | | Descriptor: | Kynurenine aminotransferase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Han, Q, Gao, Y.G, Robinson, H, Li, J. | | Deposit date: | 2007-09-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insight into the mechanism of substrate specificity of aedes kynurenine aminotransferase.
Biochemistry, 47, 2008
|
|
2RGJ
 
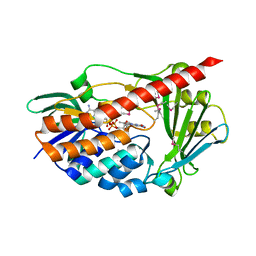 | | Crystal structure of flavin-containing monooxygenase PhzS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase | | Authors: | Ladner, J.E, Parsons, J.F, Greenhagen, B.T, Robinson, H. | | Deposit date: | 2007-10-03 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Pyocyanin Biosynthetic Protein PhzS.
Biochemistry, 47, 2008
|
|
2R5E
 
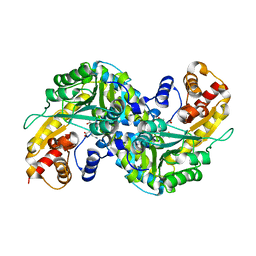 | | Aedes kynurenine aminotransferase in complex with glutamine | | Descriptor: | Kynurenine aminotransferase, N~2~-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-GLUTAMINE | | Authors: | Han, Q, Gao, Y.G, Robinson, H, Li, J. | | Deposit date: | 2007-09-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insight into the mechanism of substrate specificity of aedes kynurenine aminotransferase.
Biochemistry, 47, 2008
|
|
3RBH
 
 | | Structure of alginate export protein AlgE from Pseudomonas aeruginosa | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, Alginate production protein AlgE, ... | | Authors: | Whitney, J.C, Hay, I.D, Li, C, Eckford, P.D, Robinson, H, Amaya, M.F, Wood, L.F, Ohman, D.E, Bear, C.E, Rehm, B.H, Howell, P.L. | | Deposit date: | 2011-03-29 | | Release date: | 2011-07-27 | | Last modified: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for alginate secretion across the bacterial outer membrane.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4MBQ
 
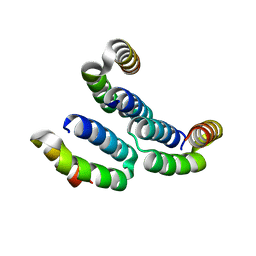 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Robinson, H, Wolfram, F, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
3IN0
 
 | |
3IN2
 
 | |
4NK8
 
 | |
4NK6
 
 | |
3JT2
 
 | |
3K9Z
 
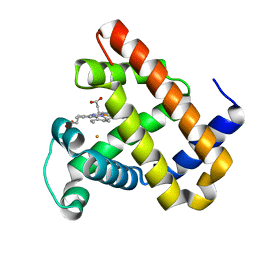 | | Rational Design of a Structural and Functional Nitric Oxide Reductase | | Descriptor: | FE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yeung, N, Lin, Y.-W, Gao, Y.-G, Zhao, X, Russell, B.S, Lei, L, Miner, K.D, Robinson, H, Lu, Y. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of a structural and functional nitric oxide reductase.
Nature, 462, 2009
|
|
4OZX
 
 | |
4O8V
 
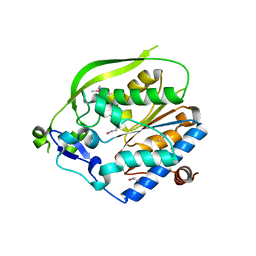 | | O-Acetyltransferase Domain of Pseudomonas putida AlgJ | | Descriptor: | Alginate biosynthesis protein AlgJ | | Authors: | Ricer, T, Little, D.J, Whitney, J.C, Robinson, H, Howell, P.L. | | Deposit date: | 2013-12-30 | | Release date: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (1.815 Å) | | Cite: | P. aeruginosa SGNH Hydrolase-Like Proteins AlgJ and AlgX Have Similar Topology but Separate and Distinct Roles in Alginate Acetylation.
Plos Pathog., 10, 2014
|
|
3JTB
 
 | |
4OZZ
 
 | |
4OZY
 
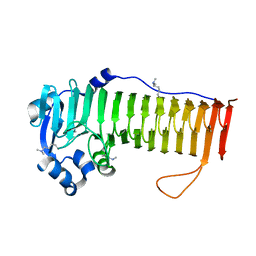 | |
4P7O
 
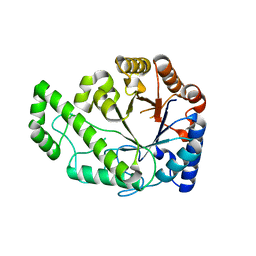 | | Structure of Escherichia coli PgaB C-terminal domain, P1 crystal form | | Descriptor: | Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7Q
 
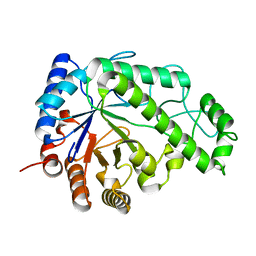 | | Structure of Escherichia coli PgaB C-terminal domain in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7R
 
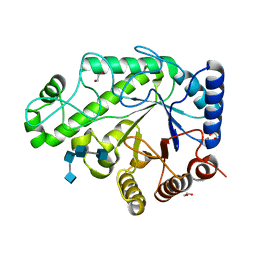 | | Structure of Escherichia coli PgaB C-terminal domain in complex with a poly-beta-1,6-N-acetyl-D-glucosamine (PNAG) hexamer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7N
 
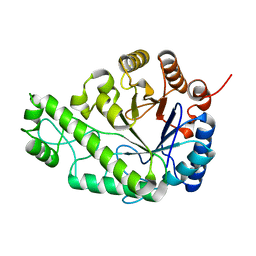 | | Structure of Escherichia coli PgaB C-terminal domain in complex with glucosamine | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7L
 
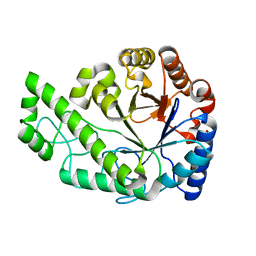 | | Structure of Escherichia coli PgaB C-terminal domain, P212121 crystal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
