1O7I
 
 | |
1KCZ
 
 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Mg-complex. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
1KD0
 
 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Apo-structure. | | Descriptor: | 1,2-ETHANEDIOL, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
3OHL
 
 | |
3OHO
 
 | |
2BI8
 
 | |
2ARD
 
 | | The structure of tryptophan 7-halogenase (PrnA) suggests a mechanism for regioselective chlorination | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, tryptophan halogenase PrnA | | Authors: | Dong, C, Flecks, S, Unversucht, S, Haupt, C, Van Pee, K.H, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2005-08-19 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Science, 309, 2005
|
|
2APG
 
 | | The structure of tryptophan 7-halogenase (PrnA)suggests a mechanism for regioselective chlorination | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Dong, C, Flecks, S, Unversucht, S, Haupt, C, Van Pee, K.H, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2005-08-16 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Science, 309, 2005
|
|
1I8T
 
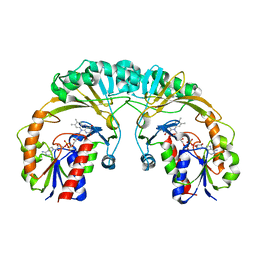 | | STRUCTURE OF UDP-GALACTOPYRANOSE MUTASE FROM E.COLI | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Sanders, D.A.R, Staines, A.G, McMahon, S.A, McNeil, M.R, Whitfield, C, Naismith, J.H. | | Deposit date: | 2001-03-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | UDP-galactopyranose mutase has a novel structure and mechanism.
Nat.Struct.Biol., 8, 2001
|
|
1WAM
 
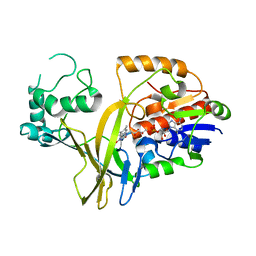 | |
1WBH
 
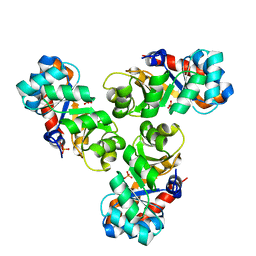 | |
1WA3
 
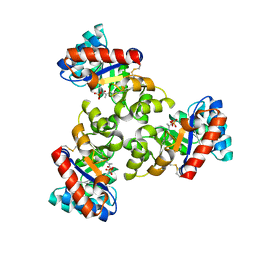 | | Mechanism of the Class I KDPG aldolase | | Descriptor: | 2-KETO-3-DEOXY-6-PHOSPHOGLUCONATE ALDOLASE, PYRUVIC ACID, SULFATE ION | | Authors: | Fullerton, S.W.B, Griffiths, J.S, Merkel, A.B, Wymer, N.J, Hutchins, M.J, Fierke, C.A, Toone, E.J, Naismith, J.H. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of the Class I Kdpg Aldolase.
Bioorg.Med.Chem., 14, 2006
|
|
1WAU
 
 | |
1G0R
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). THYMIDINE/GLUCOSE-1-PHOSPHATE COMPLEX. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION, ... | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G23
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). GLUCOSE-1-PHOSPHATE COMPLEX. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-16 | | Release date: | 2000-12-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1FXO
 
 | |
1FZW
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). APO ENZYME. | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-04 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G1A
 
 | | THE CRYSTAL STRUCTURE OF DTDP-D-GLUCOSE 4,6-DEHYDRATASE (RMLB)FROM SALMONELLA ENTERICA SEROVAR TYPHIMURIUM | | Descriptor: | DTDP-D-GLUCOSE 4,6-DEHYDRATASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Allard, S.T.M, Giraud, M.-F, Whitfield, C, Graninger, M, Messner, P, Naismith, J.H. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The crystal structure of dTDP-D-Glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium, the second enzyme in the dTDP-l-rhamnose pathway.
J.Mol.Biol., 307, 2001
|
|
1G2V
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TTP COMPLEX. | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G3L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-L-RHAMNOSE COMPLEX. | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1RTV
 
 | | RmlC (dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase) crystal structure from Pseudomonas aeruginosa, apo structure | | Descriptor: | S,R MESO-TARTARIC ACID, dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Dong, C.J, Naismith, J.H. | | Deposit date: | 2003-12-10 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RmlC (dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase) crystal structure from Pseudomonas aeruginosa, apo structure
TO BE PUBLISHED
|
|
2X3F
 
 | | Crystal Structure of the Methicillin-Resistant Staphylococcus aureus Sar2676, a Pantothenate Synthetase. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PANTHOTHENATE SYNTHETASE, SULFATE ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
7BKA
 
 | |
7BHJ
 
 | |
2W82
 
 | | The structure of ArdA | | Descriptor: | ORF18 | | Authors: | McMahon, S.A, Roberts, G.A, Carter, L.G, Cooper, L.P, Liu, H, White, J.H, Johnson, K.A, Sanghvi, B, Oke, M, Walkinshaw, M.D, Blakely, G, Naismith, J.H, Dryden, D.T.F. | | Deposit date: | 2009-01-08 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Extensive DNA Mimicry by the Arda Anti-Restriction Protein and its Role in the Spread of Antibiotic Resistance.
Nucleic Acids Res., 37, 2009
|
|
