4ZO4
 
 | | Dephospho-CoA kinase from Campylobacter jejuni. | | Descriptor: | BETA-MERCAPTOETHANOL, Dephospho-CoA kinase | | Authors: | Osipiuk, J, Zhou, M, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-06 | | Release date: | 2015-05-13 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Dephospho-CoA kinase from Campylobacter jejuni.
to be published
|
|
4PZL
 
 | | The crystal structure of adenylate kinase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Adenylate kinase, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-31 | | Release date: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of adenylate kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
2JWQ
 
 | | G-quadruplex recognition by quinacridines: a SAR, NMR and Biological study | | Descriptor: | DNA (5'-D(*DTP*DTP*DAP*DGP*DGP*DGP*DT)-3'), N,N'-(dibenzo[b,j][1,7]phenanthroline-2,10-diyldimethanediyl)dipropan-1-amine | | Authors: | Hounsou, C, Guittat, L, Monchaud, D, Jourdan, M, Saettel, N, Mergny, J.L, Teulade-Fichou, M. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | G-Quadruplex Recognition by Quinacridines: a SAR, NMR, and Biological Study
ChemMedChem, 2, 2007
|
|
3CWC
 
 | | Crystal structure of putative glycerate kinase 2 from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative glycerate kinase 2 | | Authors: | Osipiuk, J, Zhou, M, Holzle, D, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-08 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | X-ray crystal structure of putative glycerate kinase 2 from Salmonella typhimurium LT2.
To be Published
|
|
4R7O
 
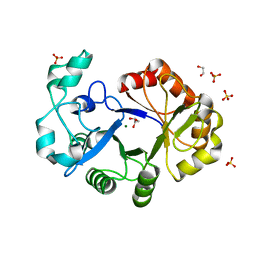 | | Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.534 Å) | | Cite: | Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci
To be Published
|
|
4R7R
 
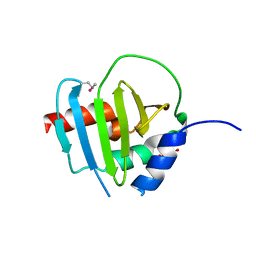 | | Crystal Structure of Putative Lipoprotein from Clostridium perfringens | | Descriptor: | GLYCEROL, Putative lipoprotein | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Crystal Structure of Putative Lipoprotein from Clostridium perfringens
To be Published
|
|
4R7Q
 
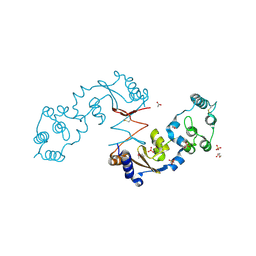 | | The structure of a sensor domain of a histidine kinase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 203, 2021
|
|
3FX3
 
 | | Structure of a putative cAMP-binding regulatory protein from Silicibacter pomeroyi DSS-3 | | Descriptor: | Cyclic nucleotide-binding protein, GLYCEROL, PHOSPHATE ION | | Authors: | Cuff, M.E, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-24 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a putative cAMP-binding regulatory protein from Silicibacter pomeroyi DSS-3
TO BE PUBLISHED
|
|
4TQ5
 
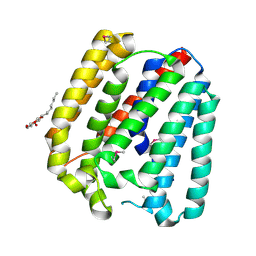 | | Structure of a UbiA homolog from Archaeoglobus fulgidus | | Descriptor: | octyl beta-D-glucopyranoside, prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2023 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
4TQ6
 
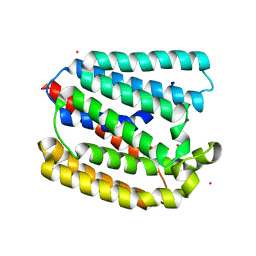 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to Cd2+ | | Descriptor: | CADMIUM ION, prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0678 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
5CJJ
 
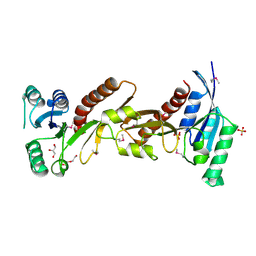 | | The crystal structure of phosphoribosylglycinamide formyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-14 | | Release date: | 2015-07-29 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The crystal structure of phosphoribosylglycinamide formyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168
To Be Published
|
|
3FWX
 
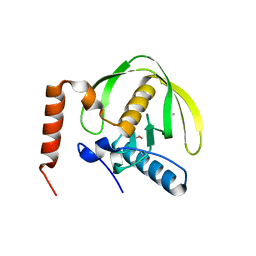 | | The crystal structure of the peptide deformylase from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | Peptide deformylase, ZINC ION | | Authors: | Zhang, R, Zhou, M, Stam, J, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the peptide deformylase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
4TQ4
 
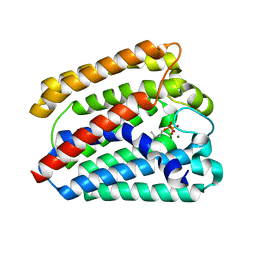 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to DMAPP and Mg2+ | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, MAGNESIUM ION, prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5025 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
3G48
 
 | | Crystal structure of chaperone CsaA form Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, Chaperone CsaA, GLYCEROL, ... | | Authors: | Nocek, B, Zhou, M, Stam, J, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-03 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of chaperone CsaA form Bacillus anthracis str. Ames
TO BE PUBLISHED
|
|
4S1N
 
 | | The crystal structure of phosphoribosylglycinamide formyltransferase from Streptococcus pneumoniae TIGR4 | | Descriptor: | CHLORIDE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-14 | | Release date: | 2015-01-28 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of phosphoribosylglycinamide formyltransferase from Streptococcus pneumoniae TIGR4
To be Published
|
|
4TQ3
 
 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to GPP and Mg2+ | | Descriptor: | GERANYL DIPHOSPHATE, MAGNESIUM ION, Prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4076 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
3GBX
 
 | | Serine hydroxymethyltransferase from Salmonella typhimurium | | Descriptor: | ACETATE ION, Serine hydroxymethyltransferase | | Authors: | Osipiuk, J, Nocek, B, Zhou, M, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of serine hydroxymethyltransferase from Salmonella typhimurium.
To be Published
|
|
4RWE
 
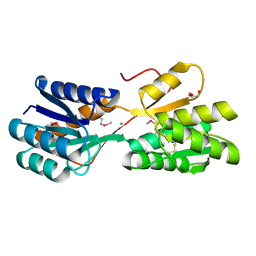 | | The crystal structure of a sugar-binding transport protein from Yersinia pestis CO92 | | Descriptor: | CHLORIDE ION, GLYCEROL, Sugar-binding transport protein | | Authors: | Tan, K, Zhou, M, Clancy, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-03 | | Release date: | 2014-12-31 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of a sugar-binding transport protein from Yersinia pestis CO92
To be Published
|
|
3CRJ
 
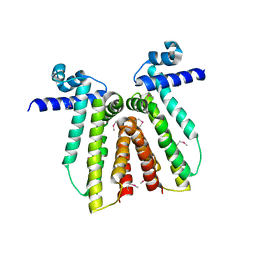 | |
3ERM
 
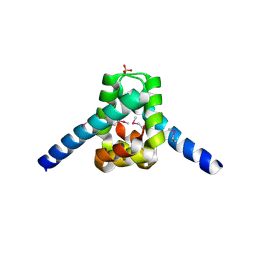 | |
3GVD
 
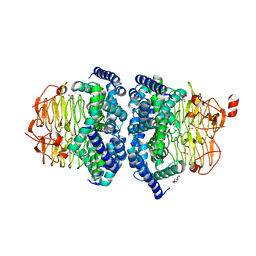 | | Crystal Structure of Serine Acetyltransferase CysE from Yersinia pestis | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETIC ACID, CYSTEINE, ... | | Authors: | Kim, Y, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-30 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Serine Acetyltransferase CysE from Yersinia pestis
To be Published
|
|
3H1S
 
 | | Crystal structure of superoxide dismutase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | FE (III) ION, GLYCEROL, Superoxide dismutase | | Authors: | Nocek, B, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-13 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of superoxide dismutase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
1DG9
 
 | | CRYSTAL STRUCTURE OF BOVINE LOW MOLECULAR WEIGHT PTPASE COMPLEXED WITH HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TYROSINE PHOSPHATASE | | Authors: | Zhang, M, Zhou, M, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 1999-11-23 | | Release date: | 1999-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of bovine low molecular weight phosphotyrosyl phosphatase complexed with the transition state analog vanadate.
Biochemistry, 36, 1997
|
|
1DDI
 
 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFITE REDUCTASE [NADPH] FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
1DDG
 
 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, SULFITE REDUCTASE (NADPH) FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
