7BTN
 
 | |
4FGX
 
 | | Crystal structure of bace1 with novel inhibitor | | Descriptor: | Beta-secretase 1, DI(HYDROXYETHYL)ETHER, Inhibitor (2R,5S,8S,12S,13S,16S,19S,22S)-16-(3-amino-3-oxopropyl)-2,13-dibenzyl-12,22-dihydroxy-8-isobutyl-19-isopropyl-3,5,17-trimethyl-4,7,10,15,18,21-hexaoxo-3,6,9,14,17,20-hexaazatricosan-1-oic acid, ... | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2012-06-05 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
7BWQ
 
 | | Structure of nonstructural protein Nsp9 from SARS-CoV-2 | | Descriptor: | Nsp9, SULFATE ION | | Authors: | Zhang, C, Chen, Y, Li, L, Su, D. | | Deposit date: | 2020-04-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.
Mol Biomed, 1, 2020
|
|
1VRY
 
 | | Second and Third Transmembrane Domains of the Alpha-1 Subunit of Human Glycine Receptor | | Descriptor: | Glycine receptor alpha-1 chain | | Authors: | Ma, D, Liu, Z, Li, L, Tang, P, Xu, Y. | | Deposit date: | 2005-07-20 | | Release date: | 2005-07-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Second and Third Transmembrane Domains of Human Glycine Receptor.
Biochemistry, 44, 2005
|
|
3HBF
 
 | | Structure of UGT78G1 complexed with myricetin and UDP | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Flavonoid 3-O-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X, Modolo, L, Li, L, Dixon, R. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids.
J.Mol.Biol., 392, 2009
|
|
6JSJ
 
 | |
3HBJ
 
 | | Structure of UGT78G1 complexed with UDP | | Descriptor: | Flavonoid 3-O-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X, Modolo, L, Li, L, Dixon, R. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids.
J.Mol.Biol., 392, 2009
|
|
4IVT
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, N-{N-[4-(acetylamino)-3,5-dichlorobenzyl]carbamimidoyl}-2-(1H-indol-1-yl)acetamide, SULFATE ION | | Authors: | Chen, T.T, Li, L, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2013-01-23 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Virtual screening and structure-based discovery of indole acylguanidines as potent beta-secretase (BACE1) inhibitors
Molecules, 18, 2013
|
|
4IVS
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, N-{N-[4-(acetylamino)-3,5-dichlorobenzyl]carbamimidoyl}-2-(6-cyano-1H-indol-1-yl)acetamide | | Authors: | Chen, T.T, Li, L, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2013-01-23 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.636 Å) | | Cite: | Virtual screening and structure-based discovery of indole acylguanidines as potent beta-secretase (BACE1) inhibitors
Molecules, 18, 2013
|
|
1Q2U
 
 | | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Huai, Q, Sun, Y, Wang, H, Chin, L.S, Li, L, Robinson, H, Ke, H. | | Deposit date: | 2003-07-26 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease
Febs Lett., 549, 2003
|
|
1Q3M
 
 | |
5XMI
 
 | | Cryo-EM Structure of the ATP-bound VPS4 mutant-E233Q hexamer (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
5XMK
 
 | | Cryo-EM structure of the ATP-bound Vps4 mutant-E233Q complex with Vta1 (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
3V69
 
 | | Filia-N crystal structure | | Descriptor: | Protein Filia | | Authors: | Wang, J, Xu, M, Zhu, K, Li, L, Liu, X. | | Deposit date: | 2011-12-19 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-terminus of FILIA Forms an Atypical KH Domain with a Unique Extension Involved in Interaction with RNA.
Plos One, 7, 2012
|
|
2LH8
 
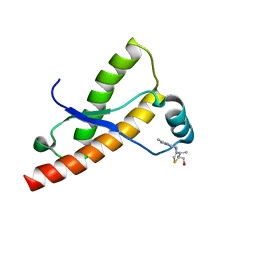 | | Syrian hamster prion protein with thiamine | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Major prion protein | | Authors: | Perez-Pineiro, R, Bjorndahl, T.C, Berjanskii, M, Hau, D, Li, L, Huang, A, Lee, R, Gibbs, E, Ladner, C, Wei Dong, Y, Abera, A, Cashman, N.R, Wishart, D. | | Deposit date: | 2011-08-05 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The prion protein binds thiamine.
Febs J., 278, 2011
|
|
2L6I
 
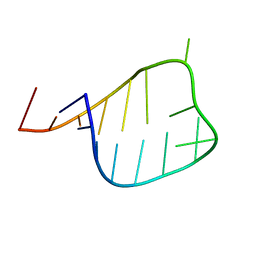 | |
3C6R
 
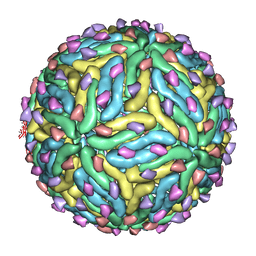 | | Low pH Immature Dengue Virus | | Descriptor: | Envelope protein, Peptide pr | | Authors: | Yu, I, Zhang, W, Holdway, H.A, Li, L, Kostyuchenko, V.A, Chipman, P.R, Kuhn, R.J, Rossmann, M.G, Chen, J. | | Deposit date: | 2008-02-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Structure of the immature dengue virus at low pH primes proteolytic maturation
Science, 319, 2008
|
|
3L8R
 
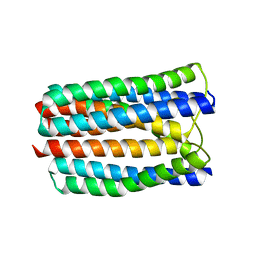 | | The crystal structure of PtcA from S. mutans | | Descriptor: | Putative PTS system, cellobiose-specific IIA component | | Authors: | Lei, J, Liu, X, Li, L. | | Deposit date: | 2010-01-03 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of PtcA from Streptococcus mutans
To be Published
|
|
3F9G
 
 | |
3F9F
 
 | |
3F9H
 
 | |
3O4O
 
 | | Crystal structure of an Interleukin-1 receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 beta, ... | | Authors: | Wang, X.Q, Wang, D.L, Zhang, S.Y, Li, L, Liu, X, Mei, K.R. | | Deposit date: | 2010-07-27 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the assembly and activation of IL-1beta with its receptors
Nat.Immunol., 11, 2010
|
|
3F9E
 
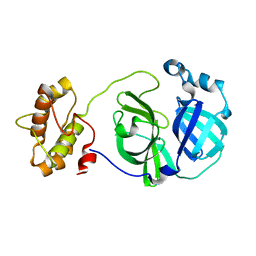 | |
3G7F
 
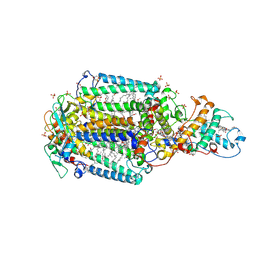 | | Crystal structure of Blastochloris viridis heterodimer mutant reaction center | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Ponomarenko, N.S, Li, L, Tereshko, V, Ismagilov, R.F, Norris Jr, J.R. | | Deposit date: | 2009-02-09 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and spectropotentiometric analysis of Blastochloris viridis heterodimer mutant reaction center
Biochim.Biophys.Acta, 1788, 2009
|
|
4OUL
 
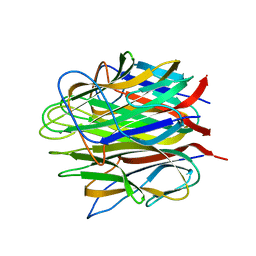 | | Crystal structure of human Caprin-2 C1q domain | | Descriptor: | CALCIUM ION, Caprin-2, GLYCEROL | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-17 | | Release date: | 2014-10-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
