3T6S
 
 | | Crystal structure of CerJ from Streptomyces tendae in Complex with CoA | | Descriptor: | COENZYME A, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-07-29 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3L89
 
 | |
3L88
 
 | | Crystal structure of the human Adenovirus type 21 fiber knob | | Descriptor: | CHLORIDE ION, Fiber protein, GLYCEROL, ... | | Authors: | Cupelli, K, Jost, M, Persson, B.D, Stehle, T. | | Deposit date: | 2009-12-30 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of adenovirus type 21 knob in complex with CD46 reveals key differences in receptor contacts among species B adenoviruses.
J.Virol., 84, 2010
|
|
3LAT
 
 | |
3T8E
 
 | | Crystal structure of CerJ from Streptomyces tendae soaked with CerviK | | Descriptor: | ACETATE ION, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-08-01 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3T5Y
 
 | | Crystal structure of CerJ from Streptomyces tendae - malonic acid covalently linked to the catalytic Cystein C116 | | Descriptor: | ACETATE ION, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3N0I
 
 | | Crystal Structure of Ad37 fiber knob in complex with GD1a oligosaccharide | | Descriptor: | Fiber, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ZINC ION | | Authors: | Nilsson, E.C, Storm, R.J, Bauer, J, Johansson, S.M.C, Lookene, A, Angstroem, J, Hedenstroem, M, Fraengsmyr, L, Rinaldi, S, Willison, H, Domelloef, F.P, Stehle, T, Arnberg, N. | | Deposit date: | 2010-05-14 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis.
NAT.MED. (N.Y.), 17, 2011
|
|
3MK4
 
 | | X-Ray structure of human PEX3 in complex with a PEX19 derived peptide | | Descriptor: | Peroxisomal biogenesis factor 19, Peroxisomal biogenesis factor 3 | | Authors: | Schmidt, F, Treiber, N, Dodt, G, Stehle, T. | | Deposit date: | 2010-04-14 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Insights into peroxisome function from the structure of PEX3 in complex with a soluble fragment of PEX19
J.Biol.Chem., 285, 2010
|
|
3O24
 
 | | Crystal structure of the brevianamide F prenyltransferase FtmPT1 from Aspergillus fumigatus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Brevianamide F prenyltransferase, CHLORIDE ION, ... | | Authors: | Jost, M, Zocher, G.E, Stehle, T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity.
J.Am.Chem.Soc., 132, 2010
|
|
3O2K
 
 | | Crystal Structure of Brevianamide F Prenyltransferase Complexed with Brevianamide F and Dimethylallyl S-thiolodiphosphate | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Brevianamide F prenyltransferase, ... | | Authors: | Jost, M, Zocher, G.E, Stehle, T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity.
J.Am.Chem.Soc., 132, 2010
|
|
4POS
 
 | |
4POR
 
 | |
4POT
 
 | |
4POQ
 
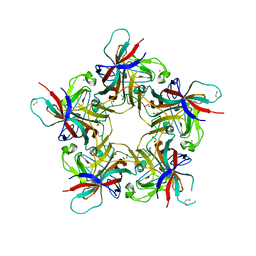 | | Structure of unliganded VP1 pentamer of Human Polyomavirus 9 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ISOPROPYL ALCOHOL, ... | | Authors: | Khan, Z.M, Stehle, T. | | Deposit date: | 2014-02-26 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and glycan microarray analysis of human polyomavirus 9 VP1 identifies N-glycolyl neuraminic acid as a receptor candidate.
J.Virol., 88, 2014
|
|
3TW5
 
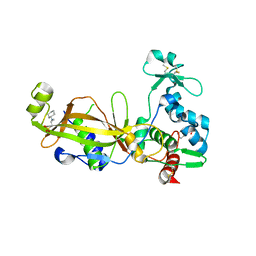 | | Crystal structure of the GP42 transglutaminase from Phytophthora sojae | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Transglutaminase elicitor | | Authors: | Reiss, K, Kirchner, E, Zocher, G, Stehle, T. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and Phylogenetic Analyses of the GP42 Transglutaminase from Phytophthora sojae Reveal an Evolutionary Relationship between Oomycetes and Marine Vibrio Bacteria.
J.Biol.Chem., 286, 2011
|
|
6Y67
 
 | |
6Y60
 
 | | Structure of Human Polyomavirus 12 VP1 in complex with 3'-Sialyllactosamine | | Descriptor: | Capsid protein VP1, N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
6Y64
 
 | | Structure of Sheep Polyomavirus VP1 in complex with 6'-Sialyllactosamine | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein VP1, GLYCEROL, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
6Y63
 
 | | Structure of Sheep Polyomavirus VP1 in complex with 3'-Sialyllactosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Capsid protein VP1, MAGNESIUM ION, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
6Y5Z
 
 | |
6Y9I
 
 | |
6Y5Y
 
 | | Structure of New Jersey Polyomavirus VP1 in complex with 3'-Sialyllactose | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
6Y65
 
 | | Structure of apo Goose Hemorrhagic Polyomavirus VP1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Capsid protein VP1, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
6Y61
 
 | |
6Y5X
 
 | |
