3BAU
 
 | |
3BAH
 
 | |
3BAO
 
 | |
3VTE
 
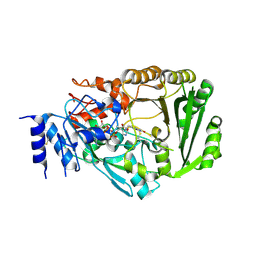 | | Crystal structure of tetrahydrocannabinolic acid synthase from Cannabis sativa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Tetrahydrocannabinolic acid synthase | | Authors: | Shoyama, Y, Tamada, T, Kurihara, K, Takeuchi, A, Taura, F, Arai, S, Blaber, M, Shoyama, Y, Morimoto, S, Kuroki, R. | | Deposit date: | 2012-05-28 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of 1-tetrahydrocannabinolic acid (THCA) synthase, the enzyme controlling the psychoactivity of Cannabis sativa
J.Mol.Biol., 423, 2012
|
|
1A80
 
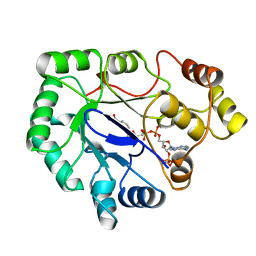 | | Native 2,5-DIKETO-D-GLUCONIC acid reductase a from CORYNBACTERIUM SP. complexed with nadph | | Descriptor: | 2,5-DIKETO-D-GLUCONIC ACID REDUCTASE A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Khurana, S, Powers, D.B, Anderson, S, Blaber, M. | | Deposit date: | 1998-03-31 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of 2,5-diketo-D-gluconic acid reductase A complexed with NADPH at 2.1-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1DYD
 
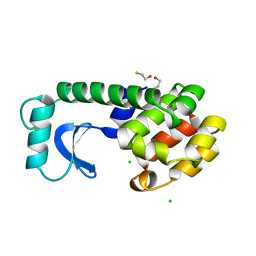 | |
1DYF
 
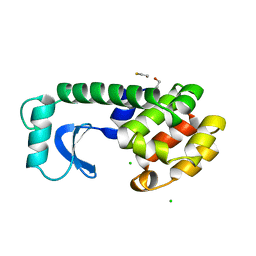 | |
1DYC
 
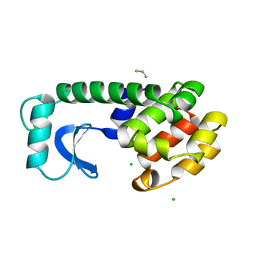 | |
1DYB
 
 | |
1DYE
 
 | |
1DYA
 
 | |
1DYG
 
 | |
4WHM
 
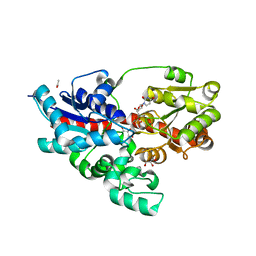 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with UDP | | Descriptor: | ACETATE ION, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
5ZCR
 
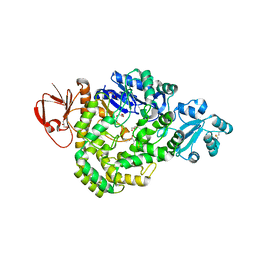 | | DSM5389 glycosyltrehalose synthase | | Descriptor: | GLYCEROL, MAGNESIUM ION, Maltooligosyl trehalose synthase | | Authors: | Tamada, T, Okazaki, N. | | Deposit date: | 2018-02-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of glycosyltrehalose synthase from Sulfolobus shibatae DSM5389
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4REN
 
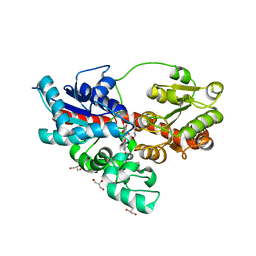 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with petunidin | | Descriptor: | 2-(3,4-dihydroxy-5-methoxyphenyl)-3,5,7-trihydroxychromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REM
 
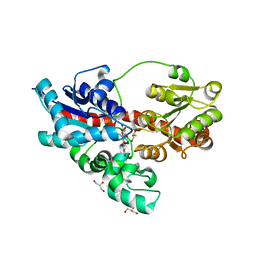 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with delphinidin | | Descriptor: | 3,5,7-trihydroxy-2-(3,4,5-trihydroxyphenyl)chromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REL
 
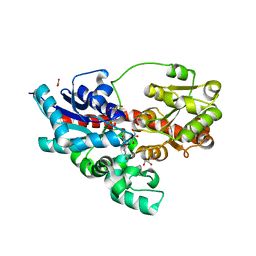 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with kaempferol | | Descriptor: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, ACETATE ION, GLYCEROL, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
1CVK
 
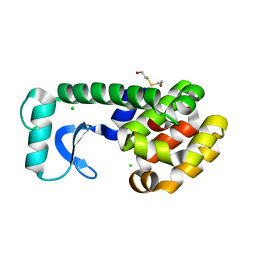 | | T4 LYSOZYME MUTANT L118A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-23 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
7XE7
 
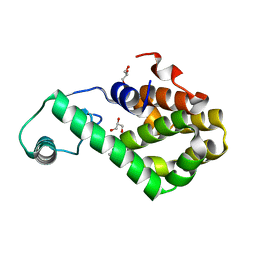 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH10 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XE6
 
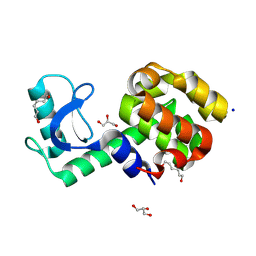 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH7 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XEA
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, DMSO 40%, and then backsoaking | | Descriptor: | CHLORIDE ION, Endolysin, GLYCEROL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XE9
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, DMSO 20% | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Endolysin, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XE5
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH4 | | Descriptor: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Tamada, T, Hiromoto, T. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
5LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
