7NIN
 
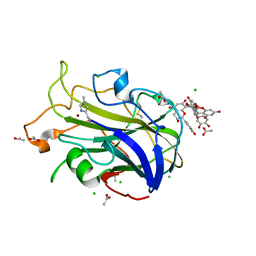 | | X-ray crystal structure of LsAA9A - CinnamtanninB1 soak | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Auxiliary activity 9, ... | | Authors: | Frandsen, K.E.H, Tokin, R, Skov, L, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of lytic polysaccharide monooxygenase by natural plant extracts.
New Phytol., 232, 2021
|
|
7NIM
 
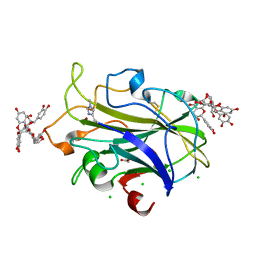 | | X-ray crystal structure of LsAA9A - cinnamon extract soak | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Auxiliary activity 9, ... | | Authors: | Frandsen, K.E.H, Tokin, R, Skov, L, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inhibition of lytic polysaccharide monooxygenase by natural plant extracts.
New Phytol., 232, 2021
|
|
1HJS
 
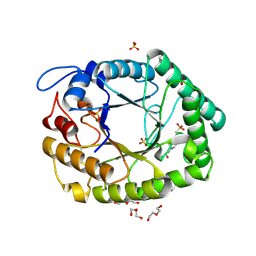 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-1,4-GALACTANASE, ... | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
1HJU
 
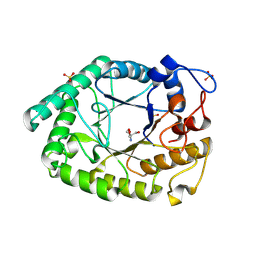 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-1,4-GALACTANASE, ... | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
1HJQ
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-1,4-GALACTANASE | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-07-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
6HSW
 
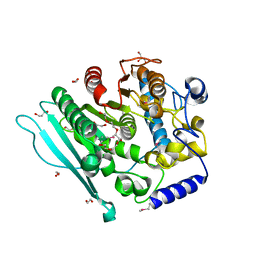 | | A CE15 glucuronoyl esterase from Teredinibacter turnerae T7901 | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Carbohydrate esterase family 15 domain protein, ... | | Authors: | Mazurkewich, S, Lo Leggio, L, Navarro Poulsen, J.C, Larsbrink, J. | | Deposit date: | 2018-10-02 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.14734387 Å) | | Cite: | Structure-function analyses reveal that a glucuronoyl esterase fromTeredinibacter turneraeinteracts with carbohydrates and aromatic compounds.
J.Biol.Chem., 294, 2019
|
|
7NN3
 
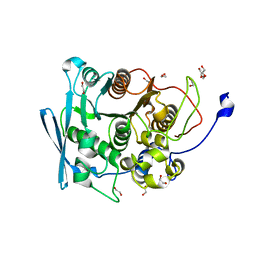 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Caldicellulosiruptor kristjansonii | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-xylanase, ... | | Authors: | Krska, D, Mazurkewich, S, Navarro Poulsen, J, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88948345 Å) | | Cite: | Structural and Functional Analysis of a Multimodular Hyperthermostable Xylanase-Glucuronoyl Esterase from Caldicellulosiruptor kristjansonii .
Biochemistry, 60, 2021
|
|
5N04
 
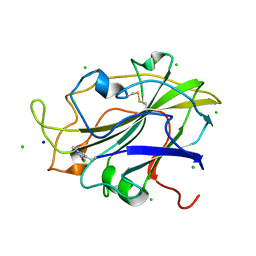 | | X-ray crystal structure of an LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Lo Leggio, L. | | Deposit date: | 2017-02-02 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Unliganded and substrate bound structures of the cellooligosaccharide active lytic polysaccharide monooxygenase LsAA9A at low pH.
Carbohydr. Res., 448, 2017
|
|
5N05
 
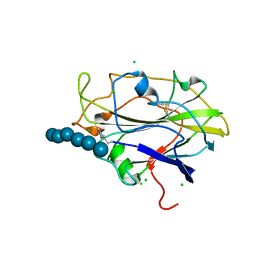 | | X-ray crystal structure of an LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Lo Leggio, L. | | Deposit date: | 2017-02-02 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Unliganded and substrate bound structures of the cellooligosaccharide active lytic polysaccharide monooxygenase LsAA9A at low pH.
Carbohydr. Res., 448, 2017
|
|
7NWQ
 
 | | A carbohydrate binding module family 9 (CBM9) from Caldicellulosiruptor kristjanssonii in complex with cellotriose | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylanase, CALCIUM ION, ... | | Authors: | Krska, D, Mazurkewich, S, Navarro Poulsen, J, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Structural and Functional Analysis of a Multimodular Hyperthermostable Xylanase-Glucuronoyl Esterase from Caldicellulosiruptor kristjansonii .
Biochemistry, 60, 2021
|
|
7NWN
 
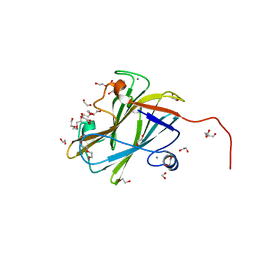 | | A carbohydrate binding module family 9 (CBM9) from Caldicellulsiruptor kristjanssonii | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylanase, CALCIUM ION, ... | | Authors: | Krska, D, Mazurkewich, S, Navarro Poulsen, J, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Functional Analysis of a Multimodular Hyperthermostable Xylanase-Glucuronoyl Esterase from Caldicellulosiruptor kristjansonii .
Biochemistry, 60, 2021
|
|
7NWO
 
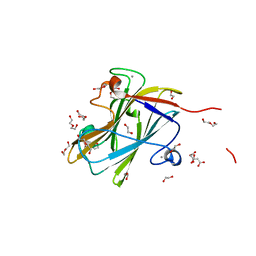 | | A carbohydrate binding module family 9 (CBM9) from Caldicellulosiruptor kristjanssonii in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylanase, CALCIUM ION, ... | | Authors: | Krska, D, Mazurkewich, S, Navarro Poulsen, J, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of a Multimodular Hyperthermostable Xylanase-Glucuronoyl Esterase from Caldicellulosiruptor kristjansonii .
Biochemistry, 60, 2021
|
|
7NWP
 
 | | A carbohydrate binding module family 9 (CBM9) from Caldicellulosiruptor kristjansonii in complex with cellobiose | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylanase, CALCIUM ION, ... | | Authors: | Krska, D, Mazurkewich, S, Navarro Poulsen, J, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.163 Å) | | Cite: | Structural and Functional Analysis of a Multimodular Hyperthermostable Xylanase-Glucuronoyl Esterase from Caldicellulosiruptor kristjansonii .
Biochemistry, 60, 2021
|
|
7OS9
 
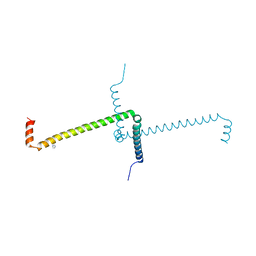 | | Crystal Structure of Domain Swapped Trp Repressor V58I Variant with purification tag | | Descriptor: | IMIDAZOLE, Trp operon repressor | | Authors: | Sprenger, J, Lawson, C.L, Lo Leggio, L, Von Wachenfeldt, C, Carey, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of Val58Ile tryptophan repressor in a domain-swapped array in the presence and absence of L-tryptophan.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7OSK
 
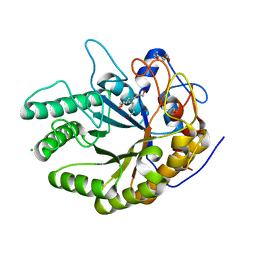 | | Ignisphaera aggregans GH53 catalytic domain | | Descriptor: | Arabinogalactan endo-1,4-beta-galactosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fredslund, F, Lo Leggio, L, Poulsen, J.C, Rasmussen, K.K, Muderspach, S, Krogh, K.B.R.M, Jensen, K. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineering the substrate binding site of the hyperthermostable archaeal endo-beta-1,4-galactanase from Ignisphaera aggregans.
Biotechnol Biofuels, 14, 2021
|
|
6ZT9
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZTA
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | Alpha-L-arabinofuranosidase | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT8
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | Alpha-L-arabinofuranosidase, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT6
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT7
 
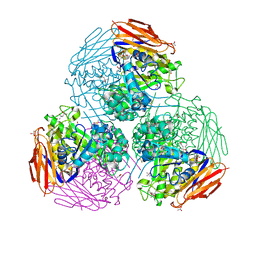 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
7BK5
 
 | | PfCopC mutant - E27A | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Muderspach, S.J, Ipsen, J, Rollan, C.H, Bertelsen, A.B, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Copper binding and reactivity at the histidine brace motif: insights from mutational analysis of the Pseudomonas fluorescens copper chaperone CopC.
Febs Lett., 595, 2021
|
|
7BK6
 
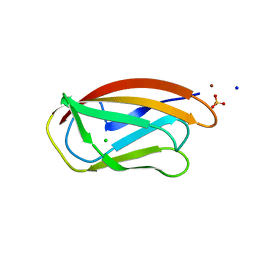 | | PfCopC mutant - D83A | | Descriptor: | ACETATE ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Muderspach, S.J, Ipsen, J, Rollan, C.H, Bertelsen, A.B, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Copper binding and reactivity at the histidine brace motif: insights from mutational analysis of the Pseudomonas fluorescens copper chaperone CopC.
Febs Lett., 595, 2021
|
|
7BK7
 
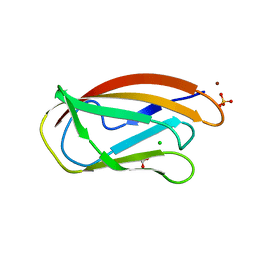 | | PfCopC mutant - D83N | | Descriptor: | ACETATE ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Muderspach, S.J, Ipsen, J, Rollan, C.H, Bertelsen, A.B, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Copper binding and reactivity at the histidine brace motif: insights from mutational analysis of the Pseudomonas fluorescens copper chaperone CopC.
Febs Lett., 595, 2021
|
|
7BF2
 
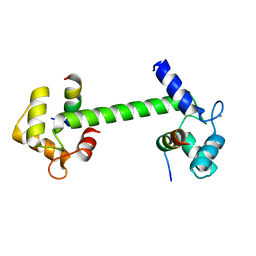 | | Ca2+-Calmodulin in complex with human muscle form creatine kinase peptide in extended 1:2 binding mode | | Descriptor: | CALCIUM ION, Calmodulin-1, Creatine kinase M-type | | Authors: | Sprenger, J, Akerfeldt, K.S, Bredfelt, J, Patel, N, Rowlett, R, Trifan, A, Vanderbeck, A, Lo Leggio, L, Snogerup Linse, S. | | Deposit date: | 2020-12-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Calmodulin complexes with brain and muscle creatine kinase peptides.
Curr Res Struct Biol, 3, 2021
|
|
7BF1
 
 | | Ca2+-Calmodulin in complex with peptide from brain-type creatine kinase in extended 1:2 binding mode | | Descriptor: | ACETYL GROUP, CALCIUM ION, Calmodulin-1, ... | | Authors: | Sprenger, J, Akerfeldt, K.S, Bredfelt, J, Patel, N, Rowlett, R, Trifan, A, Vanderbeck, A, Lo Leggio, L, Snogerup Linse, S. | | Deposit date: | 2020-12-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Calmodulin complexes with brain and muscle creatine kinase peptides.
Curr Res Struct Biol, 3, 2021
|
|
