2Q8U
 
 | |
2Q83
 
 | |
3L5O
 
 | |
3LIU
 
 | |
3M82
 
 | |
3MSW
 
 | |
3M81
 
 | |
3M83
 
 | |
3N8U
 
 | |
3NL9
 
 | |
3NPD
 
 | |
3O0F
 
 | |
2B8N
 
 | |
3CM1
 
 | |
4QDG
 
 | |
4Q20
 
 | |
4Q5K
 
 | | Crystal structure of a N-acetylmuramoyl-L-alanine amidase (BACUNI_02947) from Bacteroides uniformis ATCC 8492 at 1.30 A resolution | | Descriptor: | (2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy]propanoic acid, SODIUM ION, Uncharacterized protein | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-guided functional characterization of DUF1460 reveals a highly specific NlpC/P60 amidase family.
Structure, 22, 2014
|
|
4Q98
 
 | |
4QB7
 
 | |
4Q68
 
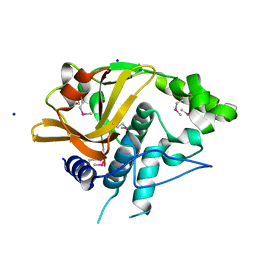 | |
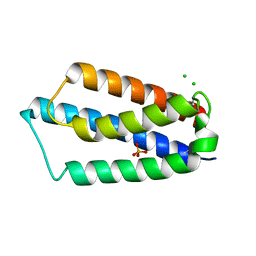
2ETS
 
 | |
4QK4
 
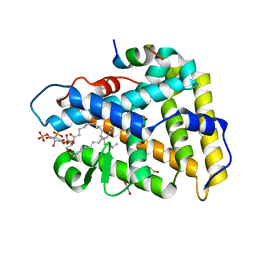 | | Crystal structure of human nuclear receptor sf-1 (nr5a1) bound to pip2 at 2.8 a resolution | | Descriptor: | (2S)-3-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2014-06-05 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2F46
 
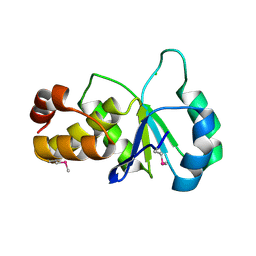 | |
2FG0
 
 | |
2FEA
 
 | |
