8CKX
 
 | |
6ZWV
 
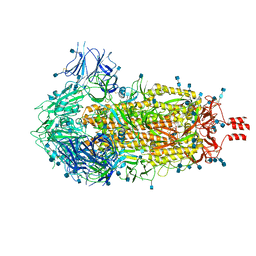 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: 3 Closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ke, Z, Qu, K, Nakane, T, Xiong, X, Cortese, M, Zila, V, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-28 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and distributions of SARS-CoV-2 spike proteins on intact virions.
Nature, 588, 2020
|
|
4D1K
 
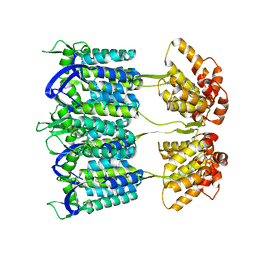 | | Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. | | Descriptor: | GAG PROTEIN | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krauesslich, H.G, Briggs, J.A.G. | | Deposit date: | 2014-05-02 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BZI
 
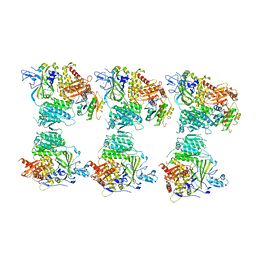 | | The structure of the COPII coat assembled on membranes | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, SAR1P, ... | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
4COP
 
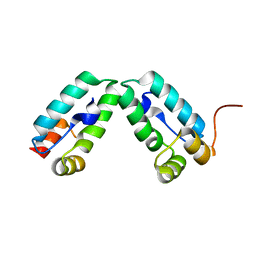 | | HIV-1 capsid C-terminal domain mutant (Y169S) | | Descriptor: | CAPSID PROTEIN P24 | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krausslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-01-29 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BZJ
 
 | | The structure of the COPII coat assembled on membranes | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
4BZK
 
 | | The structure of the COPII coat assembled on membranes | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
4COC
 
 | | HIV-1 capsid C-terminal domain mutant (Y169L) | | Descriptor: | CAPSID PROTEIN P24, SULFATE ION | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krausslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6EHM
 
 | | Model of the Ebola virus nucleocapsid subunit from recombinant virus-like particles | | Descriptor: | Membrane-associated protein VP24, Nucleoprotein | | Authors: | Wan, W, Kolesnikova, L, Clarke, M, Koehler, A, Noda, T, Becker, S, Briggs, J.A.G. | | Deposit date: | 2017-09-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structure and assembly of the Ebola virus nucleocapsid.
Nature, 551, 2017
|
|
6EHL
 
 | | Model of the Ebola virus nucleoprotein in recombinant nucleocapsid-like assemblies | | Descriptor: | Nucleoprotein | | Authors: | Wan, W, Kolesnikova, L, Clarke, M, Koehler, A, Noda, T, Becker, S, Briggs, J.A.G. | | Deposit date: | 2017-09-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure and assembly of the Ebola virus nucleocapsid.
Nature, 551, 2017
|
|
7JZT
 
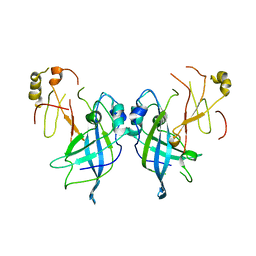 | |
7JZJ
 
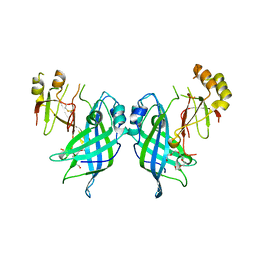 | |
5W8M
 
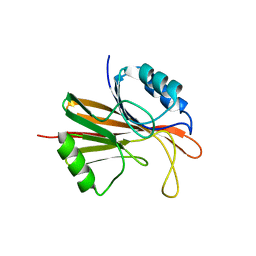 | | Crystal structure of Chaetomium thermophilum Vps29 | | Descriptor: | GLYCEROL, TRIETHYLENE GLYCOL, Vacuolar protein sorting-associated protein 29 | | Authors: | Collins, B.M, Leneva, N. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of the membrane-assembled retromer coat determined by cryo-electron tomography.
Nature, 561, 2018
|
|
5IJO
 
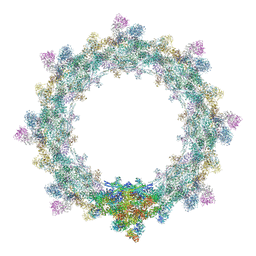 | | Alternative composite structure of the inner ring of the human nuclear pore complex (16 copies of Nup188, 16 copies of Nup205) | | Descriptor: | Nuclear pore complex protein Nup155, Nuclear pore complex protein Nup205, Nuclear pore complex protein Nup93, ... | | Authors: | Kosinski, J, Mosalaganti, S, von Appen, A, Beck, M. | | Deposit date: | 2016-03-02 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21.4 Å) | | Cite: | Molecular architecture of the inner ring scaffold of the human nuclear pore complex.
Science, 352, 2016
|
|
5IJN
 
 | | Composite structure of the inner ring of the human nuclear pore complex (32 copies of Nup205) | | Descriptor: | NUCLEAR PORE COMPLEX PROTEIN NUP155, NUCLEAR PORE COMPLEX PROTEIN NUP205, NUCLEAR PORE COMPLEX PROTEIN NUP54, ... | | Authors: | Kosinski, J, Mosalaganti, S, von Appen, A, Beck, M. | | Deposit date: | 2016-03-02 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21.4 Å) | | Cite: | Molecular architecture of the inner ring scaffold of the human nuclear pore complex.
Science, 352, 2016
|
|
5JM0
 
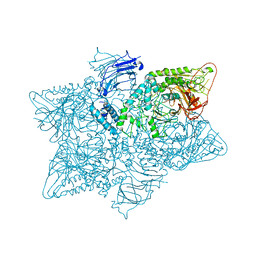 | | Structure of the S. cerevisiae alpha-mannosidase 1 | | Descriptor: | Alpha-mannosidase,Alpha-mannosidase,Alpha-mannosidase | | Authors: | Schneider, S, Kosinski, J, Jakobi, A.J, Hagen, W.J.H, Sachse, C. | | Deposit date: | 2016-04-28 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Higher-order assemblies of oligomeric cargo receptor complexes form the membrane scaffold of the Cvt vesicle.
Embo Rep., 17, 2016
|
|
5JM6
 
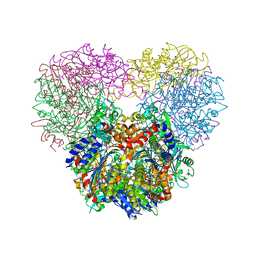 | | Structure of Chaetomium thermophilum mApe1 | | Descriptor: | Aminopeptidase-like protein, ZINC ION | | Authors: | Bertipaglia, C, Jakobi, A.J, Wilmanns, M, Sachse, C. | | Deposit date: | 2016-04-28 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Higher-order assemblies of oligomeric cargo receptor complexes form the membrane scaffold of the Cvt vesicle.
Embo Rep., 17, 2016
|
|
5JM9
 
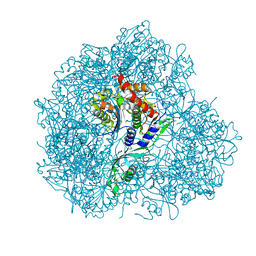 | | Structure of S. cerevesiae mApe1 dodecamer | | Descriptor: | Vacuolar aminopeptidase 1 | | Authors: | Sachse, C, Bertipaglia, C. | | Deposit date: | 2016-04-28 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Higher-order assemblies of oligomeric cargo receptor complexes form the membrane scaffold of the Cvt vesicle.
Embo Rep., 17, 2016
|
|
6EUI
 
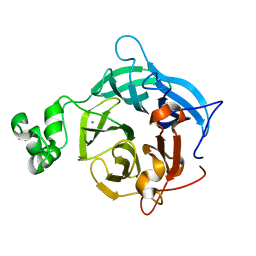 | | The GH43, Beta 1,3 Galactosidase, BT3683 with galactose | | Descriptor: | Beta-glucanase, CALCIUM ION, beta-D-galactopyranose | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUJ
 
 | | The GH43, Beta 1,3 Galactosidase, BT0265 | | Descriptor: | Beta-glucanase | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUF
 
 | | The GH43, Beta 1,3 Galactosidase, BT0265 | | Descriptor: | Beta-glucanase, alpha-L-arabinofuranose-(1-3)-[alpha-L-arabinofuranose-(1-4)][beta-D-glucopyranuronic acid-(1-6)]beta-D-galactopyranose-(1-6)-beta-D-galactopyranose, alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-6)-[alpha-L-arabinofuranose-(1-3)][alpha-L-arabinofuranose-(1-4)]beta-D-galactopyranose-(1-6)-beta-D-galactopyranose | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUH
 
 | | The GH43, Beta 1,3 Galactosidase, BT3683 with galactodeoxynojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, Beta-glucanase, CALCIUM ION | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUG
 
 | |
6EON
 
 | | Galactanase BT0290 | | Descriptor: | Beta-galactosidase, CALCIUM ION, alpha-D-galactopyranose | | Authors: | Basle, A, Munoz, J, Gilbert, H. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
5MU7
 
 | | Crystal Structure of the beta/delta-COPI Core Complex | | Descriptor: | Coatomer subunit beta, Coatomer subunit delta-like protein | | Authors: | Kopp, J, Aderhold, P, Wieland, F, Sinning, I. | | Deposit date: | 2017-01-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
