3BH8
 
 | | Crystal Structure of RQA_M Phosphopeptide Bound to HUMAN Class I MHC HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Cobbold, M, Zarling, A.L, Salim, M, Barrett-Wilt, G.A, Shabanowitz, J, Hunt, D.F, Engelhard, V.H, Willcox, B.E. | | Deposit date: | 2007-11-28 | | Release date: | 2008-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self
Nat.Immunol., 9, 2008
|
|
3BGM
 
 | | Crystal Structure of PKD2 Phosphopeptide Bound to Human Class I MHC HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Cobbold, M, Zarling, A.L, Salim, M, Barrett-Wilt, G.A, Shabanowitz, J, Hunt, D.F, Engelhard, V.H, Willcox, B.E. | | Deposit date: | 2007-11-27 | | Release date: | 2008-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self
Nat.Immunol., 9, 2008
|
|
8BT1
 
 | | YdaT transcription regulator (CII functional analog) | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Prolic-Kalinsek, M, Loris, R. | | Deposit date: | 2022-11-27 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.39788437 Å) | | Cite: | Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli O157:H7.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3BH9
 
 | | Crystal Structure of RTY Phosphopeptide Bound to Human Class I MHC HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Cobbold, M, Zarling, A.L, Salim, M, Barrett-Wilt, G.A, Shabanowitz, J, Hunt, D.F, Engelhard, V.H, Willcox, B.E. | | Deposit date: | 2007-11-28 | | Release date: | 2008-10-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self
Nat.Immunol., 9, 2008
|
|
3D25
 
 | | Crystal structure of HA-1 minor histocompatibility antigen bound to human class I MHC HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Nicholls, S, Piper, K.P, Mohammed, F, Dafforn, T.R, Tenzer, S, Salim, M, Mahendra, P, Craddock, C, van Endert, P, Schild, H, Cobbold, M, Engelhard, V.H, Moss, P.A.H, Willcox, B.E. | | Deposit date: | 2008-05-07 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Secondary anchor polymorphism in the HA-1 minor histocompatibility antigen critically affects MHC stability and TCR recognition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FRH
 
 | | Structure of the 16S rRNA methylase RmtB, P21 | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Dupechez, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-08 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
3FRI
 
 | | Structure of the 16S rRNA methylase RmtB, I222 | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Dupechez, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-08 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
331D
 
 | | CRYSTAL STRUCTURE OF D(GCGCGCG) WITH 5'-OVERHANG G'S | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*GP*CP*GP*CP*G)-3') | | Authors: | Pan, B, Ban, C, Wahl, M, Sundaralingam, M. | | Deposit date: | 1997-05-13 | | Release date: | 1997-09-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of d(GCGCGCG) with 5'-overhang G residues.
Biophys.J., 73, 1997
|
|
3FZG
 
 | | Structure of the 16S rRNA methylase ArmA | | Descriptor: | 16S rRNA methylase, S-ADENOSYLMETHIONINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-26 | | Release date: | 2009-08-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
6G5G
 
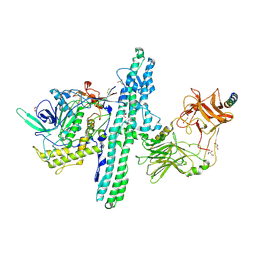 | | Crystal structure of an engineered Botulinum Neurotoxin type B mutant E1191M/S1199Y in complex with human synaptotagmin 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Masuyer, G, Elliot, M, Favre-Guilmard, C, Liu, S.M, Maignel, J, Beard, M, Carre, D, Kalinichev, M, Lezmi, S, Mir, I, Nicoleau, C, Palan, S, Perier, C, Raban, E, Dong, M, Krupp, J, Stenmark, P. | | Deposit date: | 2018-03-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered botulinum neurotoxin B with improved binding to human receptors has enhanced efficacy in preclinical models.
Sci Adv, 5, 2019
|
|
6G5F
 
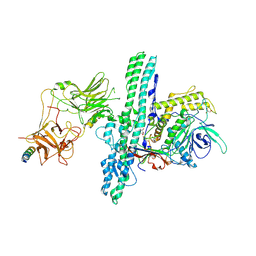 | | Crystal structure of an engineered Botulinum Neurotoxin type B mutant E1191M/S1199Y in complex with human synaptotagmin 1 | | Descriptor: | Botulinum neurotoxin type B, GLYCEROL, MALONATE ION, ... | | Authors: | Masuyer, G, Elliot, M, Favre-Guilmard, C, Liu, S.M, Maignel, J, Beard, M, Carre, D, Kalinichev, M, Lezmi, S, Mir, I, Nicoleau, C, Palan, S, Perier, C, Raban, E, Dong, M, Krupp, J, Stenmark, P. | | Deposit date: | 2018-03-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineered botulinum neurotoxin B with improved binding to human receptors has enhanced efficacy in preclinical models.
Sci Adv, 5, 2019
|
|
5EPW
 
 | | C-Terminal Domain Of Human Coronavirus Nl63 Nucleocapsid Protein | | Descriptor: | Nucleoprotein | | Authors: | Szelazek, B, Kabala, W, Kus, K, Zdzalik, M, Golik, P, Florek, D, Burmistrz, M, Pyrc, K, Dubin, G. | | Deposit date: | 2015-11-12 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Characterization of Human Coronavirus NL63 N Protein.
J. Virol., 91, 2017
|
|
6G5K
 
 | | Crystal structure of the binding domain of Botulinum Neurotoxin type B in complex with human synaptotagmin 1 | | Descriptor: | Botulinum neurotoxin type B, Synaptotagmin-1 | | Authors: | Masuyer, G, Elliot, M, Favre-Guilmard, C, Liu, S.M, Maignel, J, Beard, M, Carre, D, Kalinichev, M, Lezmi, S, Mir, I, Nicoleau, C, Palan, S, Perier, C, Raban, E, Dong, M, Krupp, J, Stenmark, P. | | Deposit date: | 2018-03-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered botulinum neurotoxin B with improved binding to human receptors has enhanced efficacy in preclinical models.
Sci Adv, 5, 2019
|
|
5H8L
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) C158S mutant in complex with putrescine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
5H8J
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) in complex with cadaverine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
5H8K
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) C158S mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
5H8I
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) in complex with N-(dihydroxymethyl)putrescine | | Descriptor: | (4-azanylbutylamino)methanediol, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
5L6S
 
 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a positive allosteric regulator beta-fructose-1,6-diphosphate (FBP) - AGPase*FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
378D
 
 | | STRUCTURE OF THE SIDE-BY-SIDE BINDING OF DISTAMYCIN TO DNA | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*GP*TP*AP*TP*AP*TP*AP*C)-3'), SODIUM ION | | Authors: | Mitra, S.N, Wahl, M.C, Sundaralingam, M. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the side-by-side binding of distamycin to d(GTATATAC)2.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
326D
 
 | | CRYSTAL STRUCTURES OF D(CM5CGGGCCM5CGG)-ORTHOGONAL FORM | | Descriptor: | DNA (5'-D(*CP*(5CM)P*GP*GP*GP*CP*CP*(5CM)P*GP*G)-3'), SPERMINE | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
317D
 
 | |
327D
 
 | |
393D
 
 | |
322D
 
 | | CRYSTAL STRUCTURES OF D(CCGGGCCM5CGG)-HEXAGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*(5CM)P*GP*G)-3') | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
394D
 
 | |
