2Q00
 
 | | Crystal structure of the P95883_SULSO protein from Sulfolobus solfataricus. NESG target SsR10. | | Descriptor: | Orf c02003 protein | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Wang, D, Owens, L, Ma, L.-C, Cunningham, K, Fang, Y, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-18 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the P95883_SULSO protein from Sulfolobus solfataricus.
To be Published
|
|
2PSB
 
 | | Crystal structure of YerB protein from Bacillus subtilis. NorthEast Structural Genomics target SR586 | | Descriptor: | YerB protein | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-04 | | Release date: | 2007-05-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of YerB protein from Bacillus subtilis.
To be Published
|
|
1DT1
 
 | | THERMUS THERMOPHILUS CYTOCHROME C552 SYNTHESIZED BY ESCHERICHIA COLI | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Fee, J.A, Chen, Y, Hill, M.J, Gomez-Moran, E, Loehr, T, Ai, J, Thony-Meyer, L, Williams, P.A, Stura, E, Sridhar, V, McRee, D.E. | | Deposit date: | 2000-01-10 | | Release date: | 2000-02-18 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integrity of thermus thermophilus cytochrome c552 synthesized by Escherichia coli cells expressing the host-specific cytochrome c maturation genes, ccmABCDEFGH: biochemical, spectral, and structural characterization of the recombinant protein.
Protein Sci., 9, 2000
|
|
3UBI
 
 | | The Absence of Tertiary Interactions in a Self-Assembled DNA Crystal Structure | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*CP*GP*TP*AP*CP*TP*CP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*AP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*CP*GP*AP*GP*TP*AP*CP*GP*AP*CP*GP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Nguyen, N, Birktoft, J.J, Sha, R, Wang, T, Zheng, J, Constantinou, P.E, Ginell, S.L, Chen, Y, Mao, C, Seeman, N.C. | | Deposit date: | 2011-10-24 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6.8046 Å) | | Cite: | The absence of tertiary interactions in a self-assembled DNA crystal structure.
J.Mol.Recognit., 25, 2012
|
|
2Q3A
 
 | | Crystal Structure of Rhesus Macaque CD8 Alpha-Alpha Homodimer | | Descriptor: | CD8 | | Authors: | Peng, H, Chen, Y, Zong, L, Gao, F, Liu, Y, Gao, G.F. | | Deposit date: | 2007-05-30 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Rhesus Macaque CD8 Homodimer Sheds the Lights on the Molecular Basis of Rhesus Macaque MHC class I with CD8 Complex
To be Published
|
|
2QGZ
 
 | | Crystal structure of a putative primosome component from Streptococcus pyogenes serotype M3. Northeast Structural Genomics target DR58 | | Descriptor: | PHOSPHATE ION, Putative primosome component | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative primosome component from Streptococcus pyogenes serotype M3.
To be Published
|
|
1SOR
 
 | | Aquaporin-0 membrane junctions reveal the structure of a closed water pore | | Descriptor: | Aquaporin-0 | | Authors: | Gonen, T, Sliz, P, Kistler, J, Cheng, Y, Walz, T. | | Deposit date: | 2004-03-15 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | Aquaporin-0 membrane junctions reveal the structure of a closed water pore
Nature, 429, 2004
|
|
3PUT
 
 | | Crystal Structure of the CERT START domain (mutant V151E) from Rhizobium etli at the resolution 1.8A, Northeast Structural Genomics Consortium Target ReR239. | | Descriptor: | HEXANE-1,6-DIOL, Hypothetical conserved protein | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Crystal Structure of the CERT START domain (mutant V151E) from Rhizobium etli at the resolution 1.8A, Northeast Structural Genomics Consortium Target ReR239.
To be Published
|
|
3LD7
 
 | | Crystal structure of the Lin0431 protein from Listeria innocua, Northeast Structural Genomics Consortium Target LkR112 | | Descriptor: | Lin0431 protein | | Authors: | Vorobiev, S, Chen, Y, Lee, D, Patel, D.J, Ciccosanti, C, Sahdev, S, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-12 | | Release date: | 2010-01-26 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Crystal structure of the Lin0431 protein from Listeria innocua
To be Published
|
|
3LD0
 
 | | Crystal structure of B.licheniformis Anti-TRAP protein, an antagonist of TRAP-RNA interactions | | Descriptor: | Inhibitor of TRAP, regulated by T-BOX (Trp) sequence RtpA, MAGNESIUM ION, ... | | Authors: | Shevtsov, M.B, Chen, Y, Isupov, M.N, Gollnick, P, Antson, A.A. | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers.
J.Struct.Biol., 170, 2010
|
|
3LRQ
 
 | | Crystal structure of the U-box domain of human ubiquitin-protein ligase (E3), NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR4604D. | | Descriptor: | E3 ubiquitin-protein ligase TRIM37, ZINC ION | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-11 | | Release date: | 2010-03-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | E3 UBIQUITIN-PROTEIN LIGASE RING DOMAIN FROM HUMAN TRIPARTITE MOTIF-CONTAINING PROTEIN 37 (TRIM37), NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR4604D.
To be Published
|
|
3L9E
 
 | | Crystal structures of holo and Cu-deficient Cu/ZnSOD from the silkworm Bombyx mori and the implications in Amyotrophic lateral sclerosis | | Descriptor: | Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Zhang, N.-N, He, Y.-X, Li, W.-F, Zhao, F, Yan, L.-F, Zhang, G.-Z, Teng, Y.-B, Yu, J, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2010-01-05 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of holo and Cu-deficient Cu/Zn-SOD from the silkworm Bombyx mori and the implications in amyotrophic lateral sclerosis
Proteins, 78, 2010
|
|
3LYX
 
 | | Crystal structure of the PAS domain of the protein CPS_1291 from Colwellia psychrerythraea. Northeast Structural Genomics Consortium target id CsR222B | | Descriptor: | Sensory box/GGDEF domain protein | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-28 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the PAS domain of the protein CPS_1291 from Colwellia psychrerythraea. Northeast Structural Genomics Consortium target id CsR222B
To be Published
|
|
3QV0
 
 | | Crystal structure of Saccharomyces cerevisiae Mam33 | | Descriptor: | Mitochondrial acidic protein MAM33 | | Authors: | Jiang, Y.L, Pu, Y.G, Ma, X.X, Chen, Y, Zhou, C.Z. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures and putative interface of Saccharomyces cerevisiae mitochondrial matrix proteins Mmf1 and Mam33.
J.Struct.Biol., 175, 2011
|
|
3NXH
 
 | | Crystal Structure of the transcriptional regulator yvhJ from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR735. | | Descriptor: | transcriptional regulator yvhJ | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-13 | | Release date: | 2010-08-04 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.584 Å) | | Cite: | Crystal Structure of the transcriptional regulator yvhJ from Bacillus subtilis.
To be Published
|
|
3PU2
 
 | | Crystal Structure of the Q3J4M4_RHOS4 protein from Rhodobacter sphaeroides. Northeast Structural Genomics Consortium Target RhR263. | | Descriptor: | uncharacterized protein | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal Structure of the Q3J4M4_RHOS4 protein from Rhodobacter sphaeroides.
To be Published
|
|
3IO4
 
 | | Huntingtin amino-terminal region with 17 Gln residues - Crystal C90 | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Huntingtin fusion protein, ZINC ION | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
5YN8
 
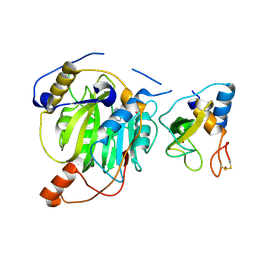 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNJ
 
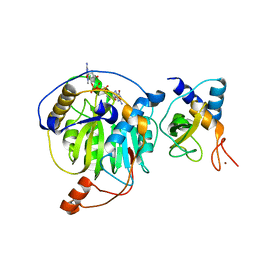 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
3QUW
 
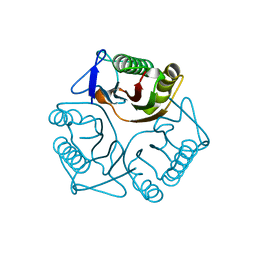 | | Crystal structure of yeast Mmf1 | | Descriptor: | Protein MMF1 | | Authors: | Jiang, Y.L, Pu, Y.G, Ma, X.X, Chen, Y, Zhou, C.Z. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures and putative interface of Saccharomyces cerevisiae mitochondrial matrix proteins Mmf1 and Mam33.
J.Struct.Biol., 175, 2011
|
|
3IO6
 
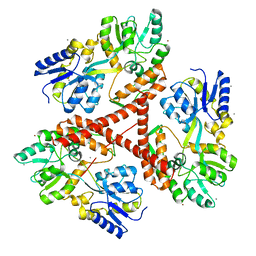 | | Huntingtin amino-terminal region with 17 Gln residues - crystal C92-a | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein, HUNTINGTIN FUSION PROTEIN, ... | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
3IMO
 
 | | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS14 | | Descriptor: | ACETATE ION, Integron cassette protein | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2009-08-11 | | Release date: | 2009-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites
Plos One, 8, 2013
|
|
1S2J
 
 | | Crystal structure of the Drosophila pattern-recognition receptor PGRP-SA | | Descriptor: | PHOSPHATE ION, Peptidoglycan recognition protein SA CG11709-PA | | Authors: | Chang, C.-I, Pili-Floury, S, Chelliah, Y, Lemaitre, B, Mengin-Lecreulx, D, Deisenhofer, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual l,d-carboxypeptidase activity.
PLOS BIOL., 2, 2004
|
|
5WQD
 
 | |
3N3Z
 
 | | Crystal structure of PDE9A (E406A) mutant in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CHLORIDE ION, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, ... | | Authors: | Hou, J, Luo, H.-B, Chen, Y, Xu, J, Zhao, R, Zou, L. | | Deposit date: | 2010-05-21 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of PDE9A (E406A) mutation in complex with IBMX
To be Published
|
|
