1YAN
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAQ
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1IOC
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME, EAEA-I56T | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Goda, S, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-02-27 | | Release date: | 2002-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Elongation in a beta-structure promotes amyloid-like fibril formation of human lysozyme.
J.Biochem., 132, 2002
|
|
1X10
 
 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192A) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1INU
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1IP7
 
 | | G129A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IP5
 
 | | G105A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IP4
 
 | | G72A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IP3
 
 | | G68A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION, SULFATE ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IP2
 
 | | G48A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IX0
 
 | | I59A-3SS human lysozyme | | Descriptor: | SODIUM ION, lysozyme | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2002-06-06 | | Release date: | 2003-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Buried water molecules contribute to the conformational stability of a protein
PROTEIN ENG., 16, 2003
|
|
1IP6
 
 | | G127A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1Z8X
 
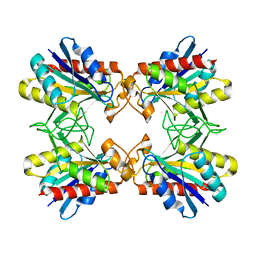 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192V) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1Z8W
 
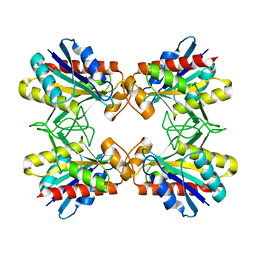 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192I) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1IP1
 
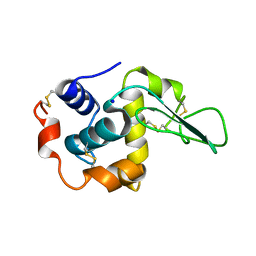 | | G37A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1Z8T
 
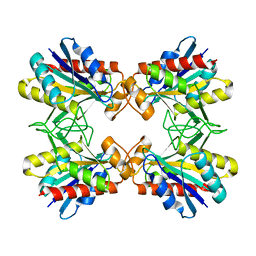 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192Q) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1V7Y
 
 | | Crystal structure of tryptophan synthase alpha-subunit from Escherichia coli at room temperature | | Descriptor: | SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Nishio, K, Morimoto, Y, Ishizuka, M, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-12-25 | | Release date: | 2005-02-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Changes in the alpha-Subunit Coupled to Binding of the beta(2)-Subunit of Tryptophan Synthase from Escherichia coli: Crystal Structure of the Tryptophan Synthase alpha-Subunit Alon
Biochemistry, 44, 2005
|
|
1X12
 
 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192D) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1V8E
 
 | |
1V8Z
 
 | | X-ray crystal structure of the Tryptophan Synthase b2 Subunit from Hyperthermophile, Pyrococcus furiosus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, Tryptophan synthase beta chain 1 | | Authors: | Hioki, Y, Ogasahara, K, Lee, S.J, Ma, J, Ishida, M, Yamagata, Y, Matsuura, Y, Ota, M, Kuramitsu, S, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of the tryptophan synthase beta subunit from the hyperthermophile Pyrococcus furiosus. Investigation of stabilization factors
Eur.J.Biochem., 271, 2004
|
|
1EQ5
 
 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-04-03 | | Release date: | 2000-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
1EQE
 
 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-04-04 | | Release date: | 2000-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
1EQ4
 
 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-04-03 | | Release date: | 2000-04-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
1V5X
 
 | |
4Y65
 
 | |
