1WLE
 
 | | Crystal Structure of mammalian mitochondrial seryl-tRNA synthetase complexed with seryl-adenylate | | Descriptor: | SERYL ADENYLATE, Seryl-tRNA synthetase | | Authors: | Chimnaronk, S, Jeppesen, M.G, Suzuki, T, Nyborg, J, Watanabe, K. | | Deposit date: | 2004-06-25 | | Release date: | 2005-09-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dual-mode recognition of noncanonical tRNAs(Ser) by seryl-tRNA synthetase in mammalian mitochondria
Embo J., 24, 2005
|
|
3WE7
 
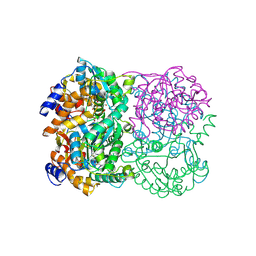 | | Crystal Structure of Diacetylchitobiose Deacetylase from Pyrococcus horikoshii | | Descriptor: | ACETIC ACID, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Mine, S, Nakamura, T, Fukuda, Y, Inoue, T, Uegaki, K, Sato, T. | | Deposit date: | 2013-07-01 | | Release date: | 2014-05-07 | | Last modified: | 2014-08-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase
Febs J., 281, 2014
|
|
3WL3
 
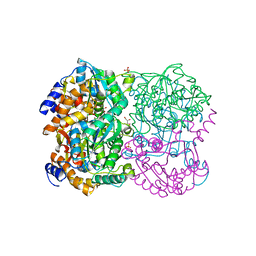 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative uncharacterized protein PH0499, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Uegaki, K. | | Deposit date: | 2013-11-07 | | Release date: | 2014-05-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase
Febs J., 281, 2014
|
|
3WL4
 
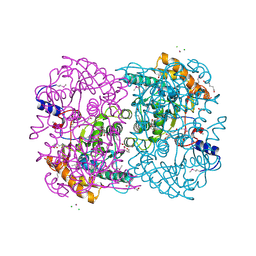 | | N,N'-diacetylchitobiose deacetylase (Se-derivative) from Pyrococcus furiosus | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Uegaki, K. | | Deposit date: | 2013-11-07 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase
Febs J., 281, 2014
|
|
5XTM
 
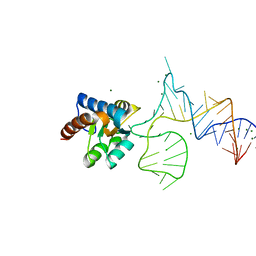 | | Crystal structure of PhoRpp38 bound to a K-turn in P12.2 helix | | Descriptor: | 50S ribosomal protein L7Ae, MAGNESIUM ION, RNA (47-MER) | | Authors: | Oshima, K, Kimura, M. | | Deposit date: | 2017-06-20 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the archaeal RNase P protein Rpp38 in complex with RNA fragments containing a K-turn motif.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3VXK
 
 | | Crystal structure of OsD14 | | Descriptor: | Dwarf 88 esterase | | Authors: | Xue, Y.-L, Miyakawa, T, Hou, F, Qin, H.-M, Tanokura, M. | | Deposit date: | 2012-09-18 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanism of strigolactone perception by DWARF14
Nat Commun, 4, 2013
|
|
5Y7M
 
 | | Crystal structure of PhoRpp38 bound to a K-turn in P12.1 helix | | Descriptor: | 50S ribosomal protein L7Ae, GUANOSINE-5'-TRIPHOSPHATE, RNA (52-MER), ... | | Authors: | Oshima, K, Kimura, M. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the archaeal RNase P protein Rpp38 in complex with RNA fragments containing a K-turn motif.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3WIO
 
 | | Crystal structure of OSD14 in complex with hydroxy D-ring | | Descriptor: | (5R)-5-hydroxy-3-methylfuran-2(5H)-one, Probable strigolactone esterase D14 | | Authors: | Xue, Y.-L, Miyakawa, T, Hou, F, Qin, H.-M, Tanokura, M. | | Deposit date: | 2013-09-22 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of strigolactone perception by DWARF14
Nat Commun, 4, 2013
|
|
2DPT
 
 | |
2DPS
 
 | |
3WO8
 
 | | Crystal structure of the beta-N-acetylglucosaminidase from Thermotoga maritima | | Descriptor: | Beta-N-acetylglucosaminidase | | Authors: | Mine, S, Kado, Y, Watanabe, M, Inoue, T, Ishikawa, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The structure of hyperthermophilic beta-N-acetylglucosaminidase reveals a novel dimer architecture associated with the active site.
Febs J., 281, 2014
|
|
3VQT
 
 | | Crystal structure analysis of the translation factor RF3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Peptide chain release factor 3 | | Authors: | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
3VR1
 
 | | Crystal structure analysis of the translation factor RF3 | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, Peptide chain release factor 3 | | Authors: | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | Deposit date: | 2012-04-03 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
