1MQQ
 
 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-1 COMPLEXED WITH GLUCURONIC ACID | | Descriptor: | ALPHA-D-GLUCURONIDASE, GLYCEROL, alpha-D-glucopyranuronic acid | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
1K9F
 
 | | Crystal structure of a mutated family-67 alpha-D-glucuronidase (E285N) from Bacillus stearothermophilus T-6, complexed with aldotetraouronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
5OKQ
 
 | | Non-conservatively refined structure of Gan1D-WT, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in complex with 6-phospho-beta-galactose | | Descriptor: | 6-O-phosphono-beta-D-galactopyranose, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
5OKE
 
 | | Conservatively refined structure of Gan1D-E170Q, a catalytic mutant of a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in complex with cellobiose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
5OKA
 
 | | Non-conservatively refined structure of Gan1D-E170Q, a catalytic mutant of a 6-phospho-beta-galactosidase from Geobacillus stearothermophilus | | Descriptor: | GLYCEROL, IMIDAZOLE, Putative 6-phospho-beta-galactobiosidase | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
5OKR
 
 | | Conservatively refined structure of Gan1D-WT, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in complex with 6-phospho-beta-glucose | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
5OKG
 
 | | Non-conservatively refined structure of Gan1D-E170Q, a catalytic mutant of a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in complex with cellobiose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
6RKJ
 
 | | The crystal structure of AbnE, an arabino-oligosaccharide binding protein, in complex with arabinooctaose | | Descriptor: | Arabino-oligosaccharids-binding protein, CALCIUM ION, GLYCEROL, ... | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
5OK7
 
 | | Conservatively refined structure of Gan1D-E170Q, a catalytic mutant of a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus | | Descriptor: | GLYCEROL, IMIDAZOLE, Putative 6-phospho-beta-galactobiosidase | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
5OKS
 
 | | Non-conservatively refined structure of Gan1D, a 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in complex with 6-phospho-beta-glucose | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
6RKH
 
 | | The crystal structure of AbnE (Selenium derivative), an arabino-oligosaccharide binding protein, in complex with arabinohexaose | | Descriptor: | Arabino-oligosaccharids-binding protein, CALCIUM ION, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.471 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
6RKX
 
 | | The crystal structure of AbnE, an arabino-oligosaccharide binding protein, in complex with arabinopentaose | | Descriptor: | Arabino-oligosaccharids-binding protein, CALCIUM ION, GLYCEROL, ... | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
6RL2
 
 | | The crystal structure of AbnE, an arabino-oligosaccharide binding protein, in complex with arabinotetraose | | Descriptor: | Arabino-oligosaccharids-binding protein, CALCIUM ION, GLYCEROL, ... | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-05-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
6RJY
 
 | | The crystal structure of AbnE, an arabino-oligosaccharide binding protein, in complex with arabinobiose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arabino-oligosaccharids-binding protein, CALCIUM ION, ... | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-04-29 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.621 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
6RKL
 
 | | The crystal structure of AbnE, an arabino-oligosaccharide binding protein, in complex with arabinoheptaose | | Descriptor: | Arabino-oligosaccharids-binding protein, CALCIUM ION, GLYCEROL, ... | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
6RL1
 
 | | The crystal structure of AbnE, an arabino-oligosaccharide binding protein, in complex with arabinotriose | | Descriptor: | Arabino-oligosaccharids-binding protein, CALCIUM ION, GLYCEROL, ... | | Authors: | Lansky, S, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2019-05-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carbohydrate-Binding Capability and Functional Conformational Changes of AbnE, an Arabino-oligosaccharide Binding Protein.
J.Mol.Biol., 432, 2020
|
|
6SWI
 
 | |
6SWL
 
 | | The REC domain of XynC, a response regulator from Geobacillus stearothermophilus | | Descriptor: | MAGNESIUM ION, Two-component response regulator | | Authors: | Lansky, S, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2019-09-22 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The REC domain of XynC, a response regulator from Geobacillus stearothermophilus
To Be Published
|
|
6SWF
 
 | |
6SWJ
 
 | | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus (with Pt) | | Descriptor: | Histidine kinase, PLATINUM (II) ION | | Authors: | Lansky, S, Shiradski, M, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2019-09-22 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus (with Pt)
To Be Published
|
|
6SW4
 
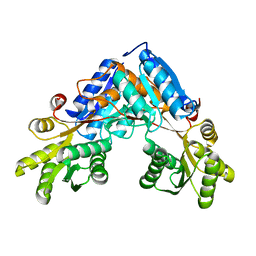 | | The structure of AraP, an arabinose binding protein from Geobacillus stearothermophilus | | Descriptor: | Arabinose binding protein | | Authors: | Lansky, S, Salama, R, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2019-09-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | The structure of AraP, an arabinose binding protein from Geobacillus stearothermophilus
To Be Published
|
|
1QW9
 
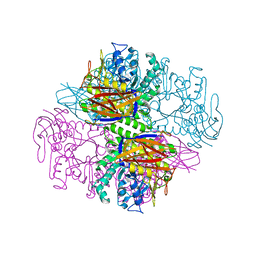 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with 4-nitrophenyl-Ara | | Descriptor: | 4-nitrophenyl alpha-L-arabinofuranoside, Alpha-L-arabinofuranosidase | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-09-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1QW8
 
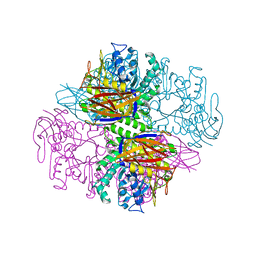 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with Ara-alpha(1,3)-Xyl | | Descriptor: | Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-09-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
6SWK
 
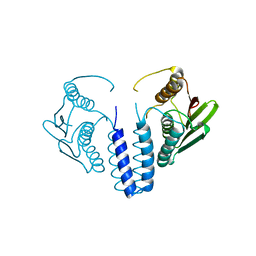 | | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus | | Descriptor: | Histidine kinase | | Authors: | Lansky, S, Shiradsky, M, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2019-09-22 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus
To Be Published
|
|
6SWB
 
 | | The REC domain of AraT, a response regulator from Geobacillus stearothermophilus | | Descriptor: | DI(HYDROXYETHYL)ETHER, Two-component response regulator | | Authors: | Lansky, S, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2019-09-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.259 Å) | | Cite: | The REC domain of AraT, a response regulator from Geobacillus stearothermophilus
To Be Published
|
|
