7W17
 
 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234E) | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VYK
 
 | |
7VYM
 
 | |
7VXH
 
 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234Q) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-12 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WSF
 
 | |
7WSE
 
 | |
7WSH
 
 | | Cryo-EM structure of SARS-CoV-2 spike receptor-binding domain in complex with sea lion ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, S, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cross-species recognition and molecular basis of SARS-CoV-2 and SARS-CoV binding to ACE2s of marine animals.
Natl Sci Rev, 9, 2022
|
|
7WSG
 
 | |
7C4Z
 
 | | Cryo-EM structure of empty Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4Y
 
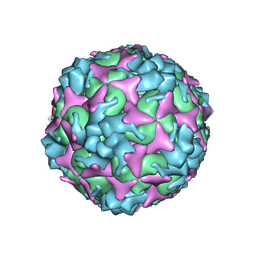 | | Cryo-EM structure of empty Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C8J
 
 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | Descriptor: | Angiotensin-converting enzyme, SARS-CoV-2 Receptor binding domain, ZINC ION | | Authors: | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7C8K
 
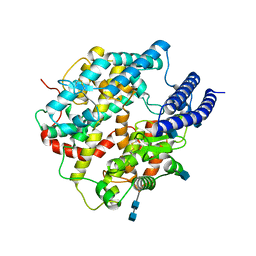 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7C4T
 
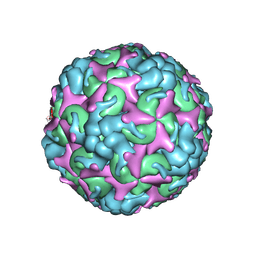 | | Cryo-EM structure of A particle Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4W
 
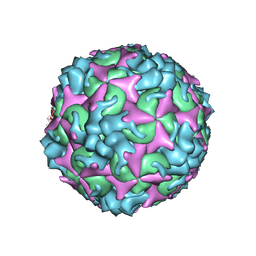 | | Cryo-EM structure of A particle Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8XS3
 
 | | Structure of MPXV B6 and D68 fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D68_heavy chain, ... | | Authors: | wu, L.L, Sun, J.Q. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Two noncompeting human neutralizing antibodies targeting MPXV B6 show protective effects against orthopoxvirus infections.
Nat Commun, 15, 2024
|
|
