8UVK
 
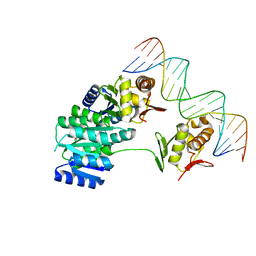 | | CosR DNA bound form II | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*T)-3'), ... | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
8UUZ
 
 | | Campylobacter jejuni CosR apo form | | Descriptor: | DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
8UVX
 
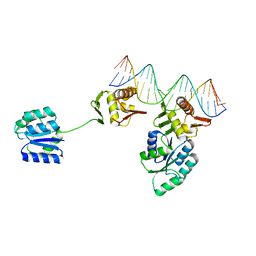 | | CosR DNA bound form I | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*TP*TP*TP*GP*GP*TP*TP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*T)-3'), DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
7F3B
 
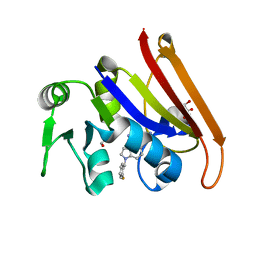 | | cocrystallization of Escherichia coli dihydrofolate reductase (DHFR) and its pyrrolo[3,2-f]quinazoline inhibitor. | | Descriptor: | 7-[(2-fluorophenyl)methyl]pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, GLYCEROL | | Authors: | Wang, H, You, X.F, Yang, X.Y, Li, Y, Hong, W. | | Deposit date: | 2021-06-16 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The discovery of 1, 3-diamino-7H-pyrrol[3, 2-f]quinazoline compounds as potent antimicrobial antifolates.
Eur.J.Med.Chem., 228, 2022
|
|
8HEV
 
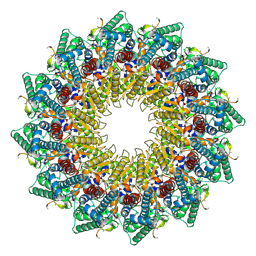 | | C12 portal in HCMV B-capsid | | Descriptor: | Portal protein, Unknown peptide | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
8HEU
 
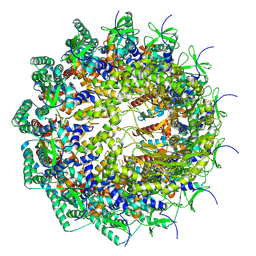 | | C12 portal in HCMV A-capsid | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
8HEY
 
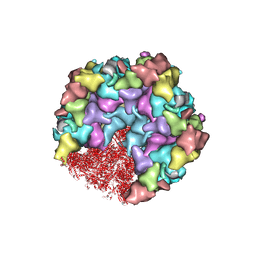 | | One CVSC-binding penton vertex in HCMV B-capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Major capsid protein, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
8HEX
 
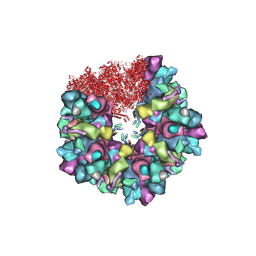 | | C5 portal vertex in HCMV B-capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Major capsid protein, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
3GX9
 
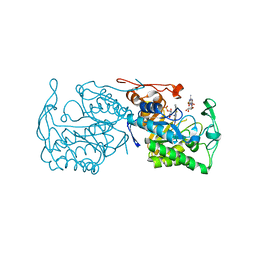 | | Structure of morphinone reductase N189A mutant in complex with tetrahydroNAD | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, Morphinone reductase | | Authors: | Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Parallel Pathways and Free-Energy Landscapes for Enzymatic Hydride Transfer Probed by Hydrostatic Pressure
Chembiochem, 10, 2009
|
|
3KFT
 
 | | Crystal structure of Pentaerythritol Tetranitrate Reductase complex with 1,4,5,6-tetrahydro NADH | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Pudney, C.R, Levy, C.W, Leys, D, Scrutton, N.S. | | Deposit date: | 2009-10-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evidence to support the hypothesis that promoting vibrations enhance the rate of an enzyme catalyzed H-tunneling reaction.
J.Am.Chem.Soc., 131, 2009
|
|
7ETM
 
 | | C6 portal vertex in the enveloped virion capsid | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
7ETJ
 
 | | C5 portal vertex in the partially-enveloped virion capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
7ET2
 
 | | Human Cytomegalovirus, C12 portal | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
7ETO
 
 | | C1 CVSC-binding penton vertex in the virion capsid of Human Cytomegalovirus | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
7ET3
 
 | | C5 portal vertex in the enveloped virion capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
4W2R
 
 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
7OO5
 
 | |
6B3W
 
 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit,Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
