5FZQ
 
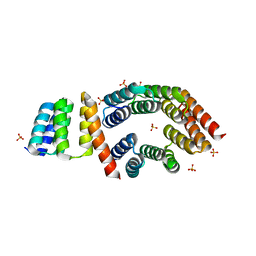 | | Designed TPR Protein M4N | | Descriptor: | DESIGNED TPR PROTEIN, SULFATE ION | | Authors: | Albrecht, R, Zhu, H, Hartmann, M.D. | | Deposit date: | 2016-03-15 | | Release date: | 2016-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Origin of a folded repeat protein from an intrinsically disordered ancestor.
Elife, 5, 2016
|
|
5FZS
 
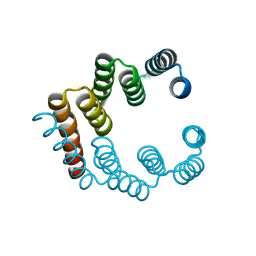 | |
2Y20
 
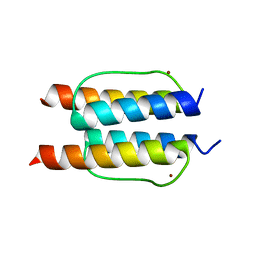 | | The mechanisms of HAMP-mediated signaling in transmembrane receptors - the A291I mutant | | Descriptor: | UNCHARACTERIZED PROTEIN, ZINC ION | | Authors: | Zeth, K, Ferris, H.U, Hulko, M, Lupas, A.N. | | Deposit date: | 2010-12-12 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanisms of Hamp-Mediated Signaling in Transmembrane Receptors.
Structure, 19, 2011
|
|
5FZR
 
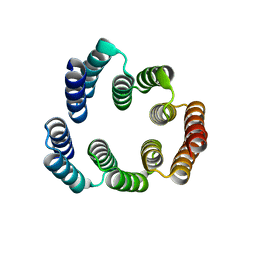 | |
2Y21
 
 | |
2WG5
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4 | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2Y0Q
 
 | |
2Y0T
 
 | |
2WG6
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4, P61A Mutant | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2M3X
 
 | |
6R60
 
 | |
6RCC
 
 | | Domain C P140 Mycoplasma genitalium | | Descriptor: | Adhesin P1, CHLORIDE ION, SODIUM ION | | Authors: | Vizarraga, D, Aparicio, D, Perez, R, Illanes, R, Fita, I. | | Deposit date: | 2019-04-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Alternative conformation of the C-domain of the P140 protein from Mycoplasma genitalium.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7TLR
 
 | |
7TLM
 
 | | Structure of Atopobium parvulum SufS | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Karunakaran, G, Couture, J.F. | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of Atopobium parvulum SufS cysteine desulfurase linked to Crohn's disease.
Febs Lett., 596, 2022
|
|
7TLQ
 
 | | Structure of Atopobium parvulum SufS C375S | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Karunakaran, G, Couture, J.F. | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of Atopobium parvulum SufS cysteine desulfurase linked to Crohn's disease.
Febs Lett., 596, 2022
|
|
7TLP
 
 | | Structure of Atopobium parvulum SufS K235R | | Descriptor: | Cysteine desulfurase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Karunakaran, G, Couture, J.F. | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural analysis of Atopobium parvulum SufS cysteine desulfurase linked to Crohn's disease.
Febs Lett., 596, 2022
|
|
6RCD
 
 | | Octamer C-Domain P140 Mycoplasma genitalium. | | Descriptor: | MgPa adhesin | | Authors: | Vizarraga, D, Aparicio, D, Perez, R, Illanes, R, Fita, I. | | Deposit date: | 2019-04-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Alternative conformation of the C-domain of the P140 protein from Mycoplasma genitalium.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4HRO
 
 | |
7ZU9
 
 | |
4HRS
 
 | |
4CSO
 
 | | The structure of OrfY from Thermoproteus tenax | | Descriptor: | ORFY PROTEIN, TRANSCRIPTION FACTOR | | Authors: | Zeth, K, Hagemann, A, Siebers, B, Martin, J, Lupas, A.N. | | Deposit date: | 2014-03-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
5NP9
 
 | |
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | Descriptor: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
2YMU
 
 | |
4GN0
 
 | | De novo phasing of a Hamp-complex using an improved Arcimboldo method | | Descriptor: | Hamp domain of AF1503, MAGNESIUM ION | | Authors: | Hulko, M, Ursinus, A, Bar, K, Martin, J, Zeth, K, Lupas, A.N. | | Deposit date: | 2012-08-16 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploiting tertiary structure through local folds for crystallographic phasing.
Nat.Methods, 10, 2013
|
|
