6QGR
 
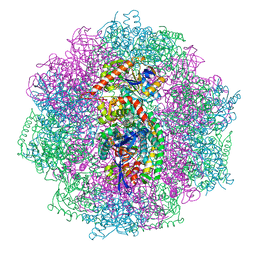 | | The F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri at the Nia-S state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit alpha, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-12 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
3DTM
 
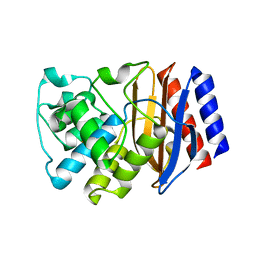 | |
7B9A
 
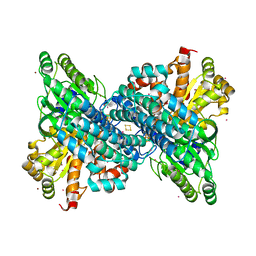 | | CooS-V with Xe-soaked | | Descriptor: | 3,5-dioxa-7-thia-1-thionia-2$l^{2},4$l^{2},6$l^{3},8$l^{2}-tetraferrabicyclo[4.2.0]octane, BROMIDE ION, Carbon monoxide dehydrogenase, ... | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Morphing [4Fe-3S-nO]-Cluster within a Carbon Monoxide Dehydrogenase Scaffold.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8ON3
 
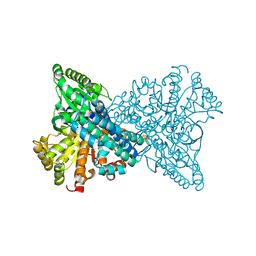 | | NI,FE-CODH -320mV + CN state : 24 h Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ON1
 
 | | NI,FE-CODH -600mV state : 4 h Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE (III) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8OMY
 
 | | NI,FE-CODH -600mV state : 35 min Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE (III) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ON0
 
 | | NI,FE-CODH -600mV state : 90 min Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE (III) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8OMX
 
 | | NI,FE-CODH -600mV state : 1 min Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(4)-NI(1)-S(5) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ON2
 
 | | NI,FE-CODH -600mV state : 24 h Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4UR0
 
 | | Crystal structure of the PCE reductive dehalogenase from S. multivorans in complex with trichloroethene | | Descriptor: | 1,1,2-trichloroethene, BENZAMIDINE, GLYCEROL, ... | | Authors: | Bommer, M, Kunze, C, Fesseler, J, Schubert, T, Diekert, G, Dobbek, H. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural Basis for Organohalide Respiration.
Science, 346, 2014
|
|
4UQU
 
 | | Crystal structure of the tetrachloroethene reductive dehalogenase from Sulfurospirillum multivorans | | Descriptor: | BENZAMIDINE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Bommer, M, Kunze, C, Fesseler, J, Schubert, T, Diekert, G, Dobbek, H. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | Structural Basis for Organohalide Respiration.
Science, 346, 2014
|
|
4UTJ
 
 | |
4UTL
 
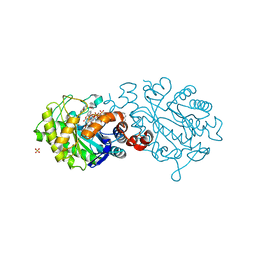 | | XenA - reduced - Y183F variant in complex with coumarin | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, COUMARIN, SULFATE ION, ... | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.229 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
4UTM
 
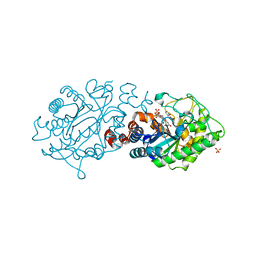 | | XenA - Reduced - Y183F variant in complex with 8-hydroxycoumarin | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 8-HYDROXYCOUMARIN, SULFATE ION, ... | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
4UR3
 
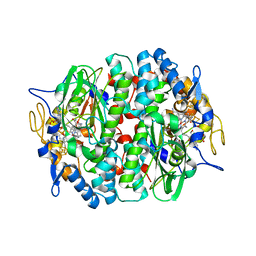 | | Crystal structure of the PCE reductive dehalogenase from S. multivorans P2(1) crystal form | | Descriptor: | IRON/SULFUR CLUSTER, NORPSEUDO-B12, TETRACHLOROETHENE REDUCTIVE DEHALOGENASE CATALYTIC SUBUNIT | | Authors: | Bommer, M, Kunze, C, Fesseler, J, Schubert, T, Diekert, G, Dobbek, H. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.235 Å) | | Cite: | Structural Basis for Organohalide Respiration.
Science, 346, 2014
|
|
4UR1
 
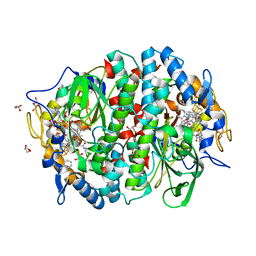 | | Crystal structure of the PCE reductive dehalogenase from S. multivorans in complex with dibromoethene | | Descriptor: | BENZAMIDINE, CIS-DIBROMOETHENE, GLYCEROL, ... | | Authors: | Bommer, M, Kunze, C, Fesseler, J, Schubert, T, Diekert, G, Dobbek, H. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural Basis for Organohalide Respiration.
Science, 346, 2014
|
|
4UR2
 
 | | Crystal structure of the PCE reductive dehalogenase from S. multivorans in complex with iodide | | Descriptor: | GLYCEROL, IODIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Bommer, M, Kunze, C, Fesseler, J, Schubert, T, Diekert, G, Dobbek, H. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural Basis for Organohalide Respiration.
Science, 346, 2014
|
|
7ZXJ
 
 | | K563A Mutant of Recombinant CODH-II | | Descriptor: | Carbon monoxide dehydrogenase 2, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Basak, Y, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | Substrate Activation at the Ni,Fe Cluster of CO Dehydrogenases: The Influence of the Protein Matrix
Acs Catalysis, 2022
|
|
7ZXL
 
 | | H93A Mutant of Recombinant CODH-II | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Activation at the Ni,Fe Cluster of CO Dehydrogenases: The Influence of the Protein Matrix
Acs Catalysis, 2022
|
|
7ZY1
 
 | | I567A Mutant of Recombinant CODH-II | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2022-05-23 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.434 Å) | | Cite: | Substrate Activation at the Ni,Fe Cluster of CO Dehydrogenases: The Influence of the Protein Matrix
Acs Catalysis, 2022
|
|
7ZX5
 
 | | I567T Mutant of Recombinant CODH-II | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.544 Å) | | Cite: | Substrate Activation at the Ni,Fe Cluster of CO Dehydrogenases: The Influence of the Protein Matrix
Acs Catalysis, 2022
|
|
7ZXC
 
 | | H96D Mutant of Recombinant CODH-II | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (III) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Substrate Activation at the Ni,Fe Cluster of CO Dehydrogenases: The Influence of the Protein Matrix
Acs Catalysis, 2022
|
|
7ZXX
 
 | | K563H Mutant of Recombinant CODH-II | | Descriptor: | Carbon monoxide dehydrogenase 2, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2022-05-23 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Substrate Activation at the Ni,Fe Cluster of CO Dehydrogenases: The Influence of the Protein Matrix
Acs Catalysis, 2022
|
|
7ZX3
 
 | | C295D Mutant of Recombinant CODH-II | | Descriptor: | Carbon monoxide dehydrogenase 2, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Substrate Activation at the Ni,Fe Cluster of CO Dehydrogenases: The Influence of the Protein Matrix
Acs Catalysis, 2022
|
|
7ZX6
 
 | | I567L Mutant of Recombinant CODH-II | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Substrate Activation at the Ni,Fe Cluster of CO Dehydrogenases: The Influence of the Protein Matrix
Acs Catalysis, 2022
|
|
