3I7I
 
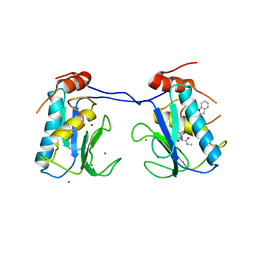 | | MMP-13 in complex with a non zinc-chelating inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, N-[4-(5-{[(1S)-1-cyclohexyl-2-(methylamino)-2-oxoethyl]carbamoyl}furan-2-yl)phenyl]-1-benzofuran-2-carboxamide, ... | | Authors: | Farrow, N.A, Margarit, S.M. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Improving potency and selectivity of a new class of non-Zn-chelating MMP-13 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4H3J
 
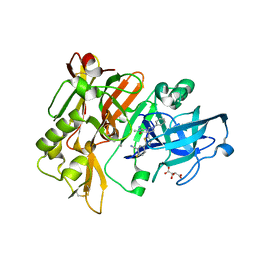 | | Structure of BACE Bound to 2-fluoro-5-(5-(2-imino-3-methyl-4-oxo-6-phenyloctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-2-yl)benzonitrile | | Descriptor: | 2-fluoro-5-{5-[(2E,4aR,7aR)-2-imino-3-methyl-4-oxo-6-phenyloctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-2-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H3F
 
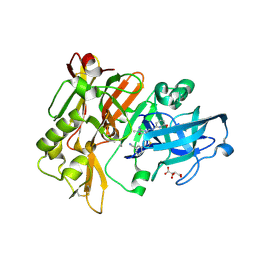 | | Structure of BACE Bound to 3-(5-((7aR)-2-imino-6-(6-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-3-yl)benzonitrile | | Descriptor: | 3-{5-[(2E,4aR,7aR)-2-imino-6-(6-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H3G
 
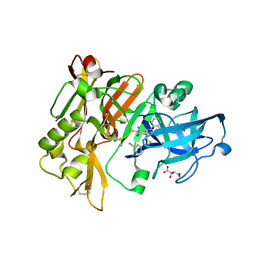 | | Structure of BACE Bound to 2-((7aR)-7a-(4-(3-cyanophenyl)thiophen-2-yl)-2-imino-3-methyl-4-oxohexahydro-1H-pyrrolo[3,4-d]pyrimidin-6(2H)-yl)nicotinonitrile | | Descriptor: | 2-{(2E,4aR,7aR)-7a-[4-(3-cyanophenyl)thiophen-2-yl]-2-imino-3-methyl-4-oxooctahydro-6H-pyrrolo[3,4-d]pyrimidin-6-yl}pyridine-3-carbonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H1E
 
 | |
4JDA
 
 | | Complex structure of abscisic acid receptor PYL3 with (-)-ABA | | Descriptor: | (2Z,4E)-5-[(1R)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Wang, G, Chen, Z. | | Deposit date: | 2013-02-24 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Insights into the Abscisic Acid Stereospecificity by the ABA Receptors PYR/PYL/RCAR
Plos One, 8, 2013
|
|
7D3Y
 
 | | Crystal structure of the osPHR2-osSPX2 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 2,Isoform 1 of Core histone macro-H2A.1 | | Authors: | Zhang, Q.X, Guan, Z.Y, Zuo, J.Q, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
7D3T
 
 | | Crystal structure of OSPHR2 in complex with DNA | | Descriptor: | DNA (5'-D(P*CP*TP*CP*GP*GP*AP*TP*AP*TP*CP*CP*TP*CP*AP*AP*G)-3'), DNA (5'-D(P*GP*CP*TP*TP*GP*AP*GP*GP*AP*TP*AP*TP*CP*CP*GP*A)-3'), Protein PHOSPHATE STARVATION RESPONSE 2 | | Authors: | Guan, Z.Y, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-20 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
5BMF
 
 | | Crystal Structure of a Theophylline binding antibody Fab fragment | | Descriptor: | 2-(5-{1-[1-(1,3-dimethyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl)-4,15-dioxo-8,11-dioxa-5,14-diazaicosan-20-yl]-3,3-dimethyl-6-sulfo-1,3-dihydro-2H-indol-2-ylidene}penta-1,3-dien-1-yl)-1-ethyl-3,3-dimethyl-3H-indolium-5-sulfonate, Fab fragment heavy chain, Fab fragment light chain | | Authors: | Bujotzek, A, Fuchs, A, Changtao, Q, Klostermann, S, Benz, J, Antes, I, Dengl, S, Hoffmann, E, Georges, G. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | MoFvAb: Modeling the Fv region of antibodies.
Mabs, 7, 2015
|
|
7FJS
 
 | | Crystal structure of T6 Fab bound to theSARS-CoV-2 RBD of B.1.351 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
4HA5
 
 | | Structure of BACE Bound to (S)-3-(5-(2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl)thiophen-3-yl)benzonitrile | | Descriptor: | 3-{5-[(2E,4S)-2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
8Y1J
 
 | |
3HGY
 
 | | Crystal Structure of CmeR Bound to Taurocholic Acid | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Routh, M.D, Yang, F. | | Deposit date: | 2009-05-14 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Structural basis for anionic ligand recognition by multidrug
binding proteins: crystal structures of CmeR-bile acid complexes
To be Published
|
|
6AVS
 
 | | Complex structure of JMJD5 and Symmetric Monomethyl-Arginine (MMA) | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, Lysine-specific demethylase 8, ZINC ION | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
1PVZ
 
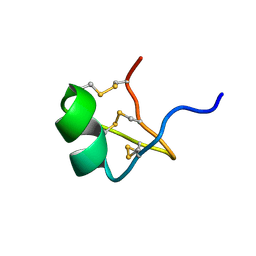 | | Solution Structure of BmP07, A Novel Potassium Channel Blocker from Scorpion Buthus martensi Karsch, 15 structures | | Descriptor: | K+ toxin-like peptide | | Authors: | Wu, H, Zhang, N, Wang, Y, Zhang, Q, Ou, L, Li, M, Hu, G. | | Deposit date: | 2003-06-29 | | Release date: | 2004-05-18 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK2, a new potassium channel blocker from the venom of chinese scorpion Buthus martensi Karsch
PROTEINS, 55, 2004
|
|
4H3I
 
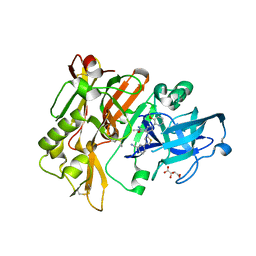 | | Structure of BACE Bound to 3-(5-((7aR)-2-imino-6-(3-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-3-yl)benzonitrile | | Descriptor: | 3-{5-[(2E,4aR,7aR)-2-imino-6-(3-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4RAY
 
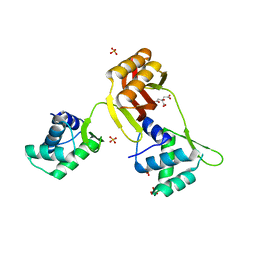 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 Apo-Fur | | Descriptor: | CITRATE ANION, DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), SULFATE ION | | Authors: | Deng, Z, Liu, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
4RB3
 
 | |
4RB0
 
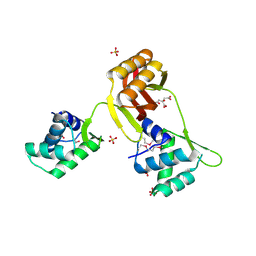 | |
1M3G
 
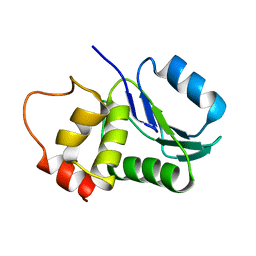 | |
4RB2
 
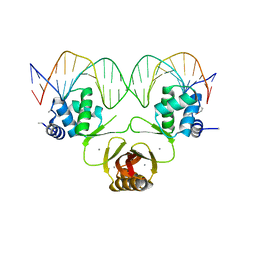 | |
4E43
 
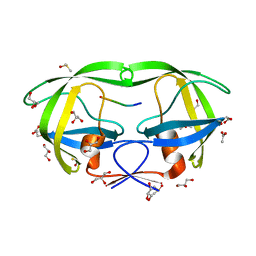 | |
8GCG
 
 | | MDM2 bound to inhibitor | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, macrocyclic peptide inhibitor | | Authors: | Silvestri, A.P, Muir, E.W, Chakka, S.K, Tripathi, S.M, Rubin, S.M, Pye, C.R, Schwochert, J.A. | | Deposit date: | 2023-03-01 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | DNA-Encoded Macrocyclic Peptide Libraries Enable the Discovery of a Neutral MDM2-p53 Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
8HJB
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA with coenzyme A | | Descriptor: | COENZYME A, TetR family transcriptional regulator | | Authors: | Liang, H, Bartlam, M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
8HC0
 
 | | Crystal structure of the extracellular domains of GPR110 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G-protein coupled receptor F1, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, F.F, Song, G.J. | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Extracellular Domains of GPR110.
J.Mol.Biol., 435, 2023
|
|
