1DN5
 
 | | SOLVATION OF THE LEFT-HANDED HEXAMER D(5BRC-G-5BRC-G-5BRC-G) IN CRYSTALS GROWN AT TWO TEMPERATURES | | Descriptor: | DNA (5'-D(*(CBR)P*GP*(CBR)P*GP*(CBR)P*G)-3') | | Authors: | Chevrier, B, Dock, A.C, Hartmann, B, Leng, M, Moras, D, Thuong, M.T, Westhof, E. | | Deposit date: | 1986-12-01 | | Release date: | 1987-04-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solvation of the left-handed hexamer d(5BrC-G-5BrC-G-5 BrC-G) in crystals grown at two temperatures.
J.Mol.Biol., 188, 1986
|
|
1DN4
 
 | | SOLVATION OF THE LEFT-HANDED HEXAMER D(5BRC-G-5BRC-G-5BRC-G) IN CRYSTALS GROWN AT TWO TEMPERATURES | | Descriptor: | DNA (5'-D(*(CBR)P*GP*(CBR)P*GP*(CBR)P*G)-3') | | Authors: | Chevrier, B, Dock, A.C, Hartmann, B, Leng, M, Moras, D, Thuong, M.T, Westhof, E. | | Deposit date: | 1986-12-01 | | Release date: | 1987-04-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Solvation of the left-handed hexamer d(5BrC-G-5BrC-G-5 BrC-G) in crystals grown at two temperatures.
J.Mol.Biol., 188, 1986
|
|
2O3V
 
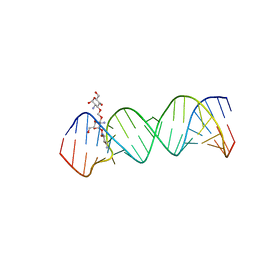 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site complexed with paromamine derivative NB33 | | Descriptor: | (2S,3R,4R,5S,6R)-3-AMINO-4-({[(2S,3R,4R,5S,6R)-3-AMINO-2-{[(1R,2R,3S,4R,6S)-4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYL]OXY}-5-HYDROXY-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-4-YL]OXY}METHOXY)-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-2,5-DIOL, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
2O3W
 
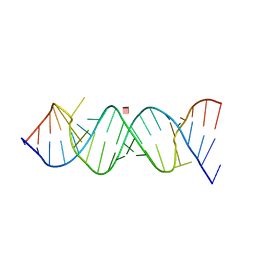 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site in presence of paromomycin | | Descriptor: | PAROMOMYCIN, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
2O3X
 
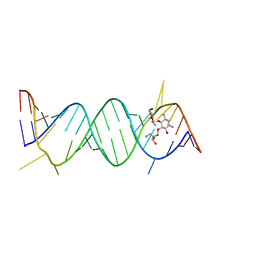 | | Crystal Structure of the Prokaryotic Ribosomal Decoding Site Complexed with Paromamine Derivative NB30 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-DIAMINO-2-[(5-AMINO-5-DEOXY-BETA-D-RIBOFURANOSYL)OXY]-3-HYDROXYCYCLOHEXYL 2-AMINO-2-DEOXY-ALPHA-D-GLUCOPYRANOSIDE, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
2O3Y
 
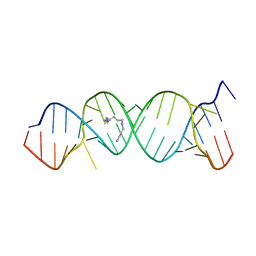 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site in Presence of Paromamine Derivative NB30 | | Descriptor: | RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3'), SPERMINE | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
2NOQ
 
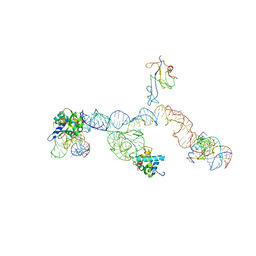 | | Structure of ribosome-bound cricket paralysis virus IRES RNA | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S5, ... | | Authors: | Schuler, M, Connell, S.R, Lescoute, A, Giesebrecht, J, Dabrowski, M, Schroeer, B, Mielke, T, Penczek, P.A, Westhof, E, Spahn, C.M.T. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structure of the ribosome-bound cricket paralysis virus IRES RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2PWT
 
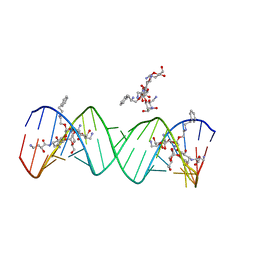 | | Crystal structure of the bacterial ribosomal decoding site complexed with aminoglycoside containing the L-HABA group | | Descriptor: | 22-mer of the ribosomal decoding site, DOUBLY FUNCTIONALIZED PAROMOMYCIN PM-II-162 | | Authors: | Kondo, J, Pachamuthu, K, Francois, B, Szychowski, J, Hanessian, S, Westhof, E. | | Deposit date: | 2007-05-13 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Bacterial Ribosomal Decoding Site Complexed with a Synthetic Doubly Functionalized Paromomycin Derivative: a New Specific Binding Mode to an A-Minor Motif Enhances in vitro Antibacterial Activity
Chemmedchem, 2, 2007
|
|
2BE0
 
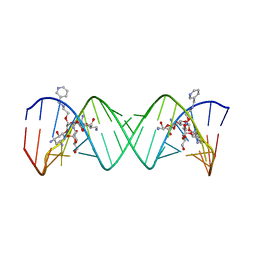 | | Complex Between Paromomycin Derivative JS5-39 and the 16S-Rrna A-Site. | | Descriptor: | (2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3R,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R,4R,5S,6R)-3-AMINO-4 ,5-DIHYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-2-(HYDROXYMETHYL)-4-(2-((R)-PIPERIDI N-3-YLMETHYLAMINO)ETHOXY)-TETRAHYDROFURAN-3-YLOXY)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Francois, B, Westhof, E. | | Deposit date: | 2005-10-21 | | Release date: | 2005-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Antibacterial aminoglycosides with a modified mode of binding to the ribosomal-RNA decoding site
ANGEW.CHEM.INT.ED.ENGL., 43, 2004
|
|
2BEE
 
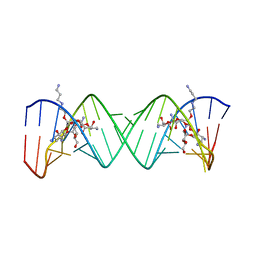 | | Complex Between Paromomycin derivative JS4 and the 16S-Rrna A Site | | Descriptor: | (2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3R,4R,5S)-4-(2-(3-AMINOPROPYLAMINO)ETHOXY)-5-((1R,2R,3S,5R,6S)-3,5-DIAM INO-2-((2S,3R,4R,5S,6R)-3-AMINO-4,5-DIHYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-2-( HYDROXYMETHYL)-TETRAHYDROFURAN-3-YLOXY)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Francois, B, Westhof, E. | | Deposit date: | 2005-10-24 | | Release date: | 2005-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Antibacterial aminoglycosides with a modified mode of binding to the ribosomal-RNA decoding site
ANGEW.CHEM.INT.ED.ENGL., 43, 2004
|
|
1VTQ
 
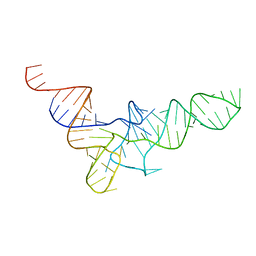 | | THREE-DIMENSIONAL STRUCTURE OF YEAST T-RNA-ASP. I. STRUCTURE DETERMINATION | | Descriptor: | T-RNA-ASP | | Authors: | Comarmond, M.B, Giege, R, Thierry, J.C, Moras, D, Fischer, J. | | Deposit date: | 1985-06-11 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-Dimensional Structure of Yeast T-RNA-ASP. I. Structure Determination
Acta Crystallogr.,Sect.B, 42, 1986
|
|
1MNX
 
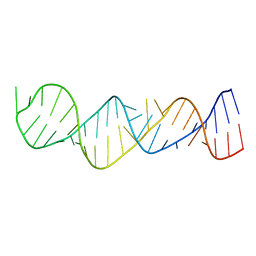 | |
5EL6
 
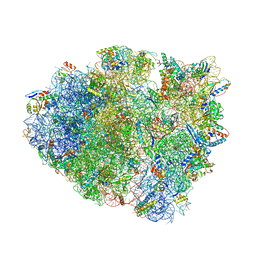 | | Structure of T. thermophilus 70S ribosome complex with mRNA and tRNALys in the A-site with a U-U mismatch in the first position and antibiotic paromomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2015-11-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
4V87
 
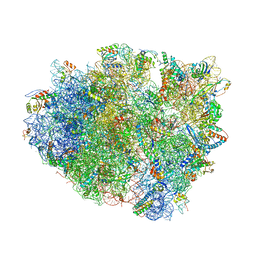 | | Crystal structure analysis of ribosomal decoding. | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Demeshkina, N, Jenner, L, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-09-20 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V8D
 
 | | Structure analysis of ribosomal decoding (cognate tRNA-tyr complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
5E81
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA and tRNALys in the A-site with wobble pair | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2015-10-13 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
5EL5
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA and tRNALys in the A-site with a U-U mismatch in the second position | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2015-11-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
6QNQ
 
 | | 70S ribosome initiation complex (IC) with experimentally assigned potassium ions | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2019-02-11 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction.
Nat Commun, 10, 2019
|
|
6QNR
 
 | | 70S ribosome elongation complex (EC) with experimentally assigned potassium ions | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2019-02-11 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction.
Nat Commun, 10, 2019
|
|
5E7K
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA and cognate tRNALys in the A-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2015-10-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
4WZD
 
 | | Complex of 70S ribosome with cognate tRNA-Tyr in the P-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-11-19 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WSM
 
 | | Complex of 70S ribosome with tRNA-Leu and mRNA with G-U mismatch in the first position in the A- and P-sites | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WRO
 
 | | Complex of 70S ribosome with tRNA-Phe and mRNA with C-A mismatch in the second position in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WQR
 
 | | Complex of 70S ribosome with tRNA-Phe and mRNA with C-A mismatch in the first position in the A-site. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-22 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WR6
 
 | | Complex of 70S ribosome with tRNA-Tyr and mRNA with A-A mismatch in the first position in the A-site. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-23 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
