3IVP
 
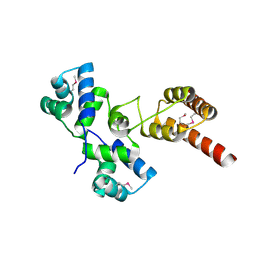 | |
5JRO
 
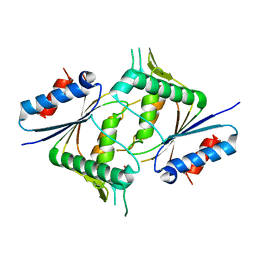 | | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form | | Descriptor: | FMN-dependent NADH-azoreductase, GLYCEROL | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-06 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form
To Be Published
|
|
5IZN
 
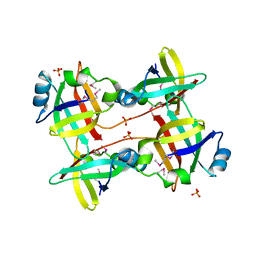 | | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6 | | Descriptor: | 50S ribosomal protein L25, PHOSPHATE ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-25 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6
To Be Published
|
|
5JQW
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, N5-carboxyaminoimidazole ribonucleotide synthase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-05 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP
To Be Published
|
|
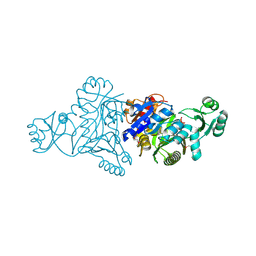
6W4B
 
 | | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 | | Descriptor: | Non-structural protein 9 | | Authors: | Tan, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of Nsp9 replicase protein of COVID-19
To Be Published
|
|
6NLP
 
 | | The crystal structure of an ABC transporter periplasmic binding protein YdcS from Escherichia coli BW25113 | | Descriptor: | 1,2-ETHANEDIOL, Bacterial extracellular solute-binding family protein, IMIDAZOLE | | Authors: | Tan, K, SKarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an ABC transporter periplasmic binding protein YdcS from Escherichia coli BW25113
To Be Published
|
|
6E4B
 
 | |
6OZV
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-16 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
6P1J
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo2 serine module | | Descriptor: | ACETATE ION, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-20 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
5JMB
 
 | |
6P4U
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module in complex with Mg and AMP | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-28 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
6OYF
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
6P3I
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module in complex with Mg | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
5JMU
 
 | | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 | | Descriptor: | ACETATE ION, MAGNESIUM ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Tan, K, Gu, M, Clancy, S, Joachimiak, A. | | Deposit date: | 2016-04-29 | | Release date: | 2016-06-29 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 (CASP target)
To Be Published
|
|
6DKH
 
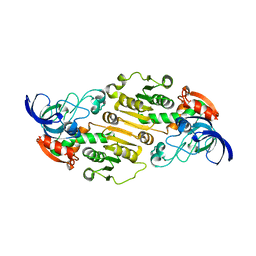 | | The crystal structure of L-idonate 5-dehydrogenase from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | L-idonate 5-dehydrogenase (NAD(P)(+)), ZINC ION | | Authors: | Tan, K, Evdokimova, E, McChesney, C, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | The crystal structure of L-idonate 5-dehydrogenase from Escherichia coli str. K-12 substr. MG1655
To Be Published
|
|
6V6N
 
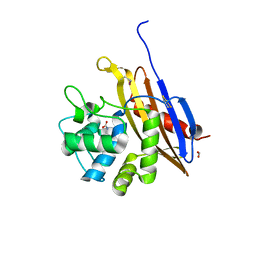 | | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens | | Descriptor: | Beta-lactamase, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-05 | | Release date: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens
To Be Published
|
|
5UFH
 
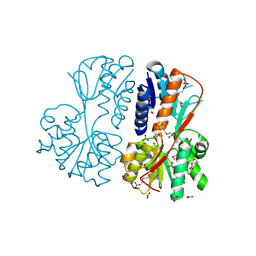 | | The crystal structure of a LacI-type transcription regulator from Bifidobacterium animalis subsp. lactis DSM 10140 | | Descriptor: | GLYCEROL, LacI-type transcriptional regulator, NITRATE ION | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of a LacI-type transcription regulator from Bifidobacterium animalis subsp. lactis DSM 10140
To Be Published
|
|
5UHJ
 
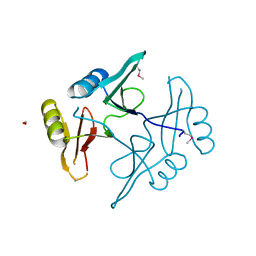 | | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234 | | Descriptor: | FORMIC ACID, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-11 | | Release date: | 2017-01-25 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234
To Be Published
|
|
1Z78
 
 | |
5UJP
 
 | | The crystal structure of a glyoxalase/bleomycin resistance protein from Streptomyces sp. CB03234 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The crystal structure of a glyoxalase/bleomycin resistance protein from Streptomyces sp. CB03234
To Be Published
|
|
1ZA4
 
 | |
6V4W
 
 | | The crystal structure of a beta-lactamase from Chitinophaga pinensis DSM 2588 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, Beta-lactamase, ... | | Authors: | Tan, K, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-02 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The crystal structure of a beta-lactamase from Chitinophaga pinensis DSM 2588
To Be Published
|
|
5UID
 
 | | The crystal structure of an aminotransferase TlmJ from Streptoalloteichus hindustanus | | Descriptor: | Aminotransferase TlmJ, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Bearden, J, Phillips Jr, G.N, Joachmiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-01 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The crystal structure of an aminotransferase TlmJ from Streptoalloteichus hindustanus.
To Be Published
|
|
5UNC
 
 | | The crystal structure of PHOSPHOENOLPYRUVATE PHOSPHOMUTASE from Streptomyces platensis subsp. rosaceus | | Descriptor: | FORMIC ACID, L(+)-TARTARIC ACID, PHOSPHOENOLPYRUVATE PHOSPHOMUTASE, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-30 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The crystal structure of PHOSPHOENOLPYRUVATE PHOSPHOMUTASE from Streptomyces platensis subsp. rosaceus
To Be Published
|
|
5USW
 
 | | The crystal structure of 7,8-dihydropteroate synthase from Vibrio fischeri ES114 | | Descriptor: | ACETATE ION, Dihydropteroate synthase, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.643 Å) | | Cite: | The crystal structure of 7,8-dihydropteroate synthase from Vibrio fischeri ES114
To Be Published
|
|
