7XGZ
 
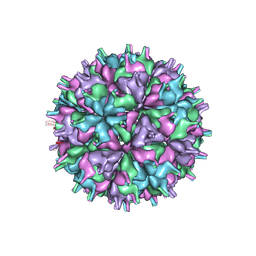 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 7.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions
Nat Commun, 14, 2023
|
|
7XPA
 
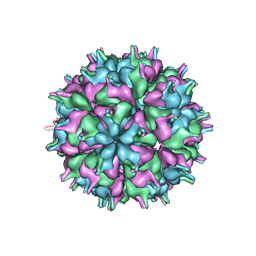 | | Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 7.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
7XPD
 
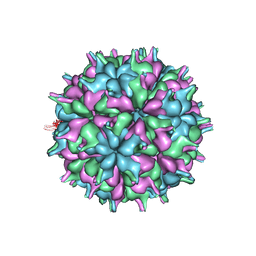 | | Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 6.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
5BUN
 
 | | Crystal structure of an antigenic outer membrane protein ST50 from Salmonella Typhi | | Descriptor: | Outer membrane protein, octyl beta-D-glucopyranoside | | Authors: | Yoshimura, M, Chuankhayan, P, Lin, C.C, Chen, N.C, Yang, M.C, Fun, H.K. | | Deposit date: | 2015-06-04 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structure of an antigenic outer-membrane protein from Salmonella Typhi suggests a potential antigenic loop and an efflux mechanism.
Sci Rep, 5, 2015
|
|
3LFI
 
 | |
5BND
 
 | | Crystal structure of the C-terminal domain of TagH | | Descriptor: | ABC transporter, ATP-binding protein | | Authors: | Chen, S.C, Chen, Y. | | Deposit date: | 2015-05-26 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | SH3-Like Motif-Containing C-terminal Domain of Staphylococcal Teichoic Acid Transporter Suggests Possible Function.
Proteins, 2016
|
|
3LF7
 
 | |
3LDK
 
 | | Crystal Structure of A. japonicus CB05 | | Descriptor: | Fructosyltransferase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Chuankhayan, P, Chen, C.J, Chaing, C.M, Hsieh, C.Y, Chen, C.D, Hsieh, Y.C. | | Deposit date: | 2010-01-13 | | Release date: | 2010-05-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Aspergillus japonicus fructosyltransferase complex with donor/acceptor substrates reveal complete sbusites in the active site for catalysis
To be Published
|
|
3LIH
 
 | |
3LIG
 
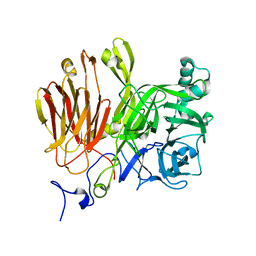 | |
3LEM
 
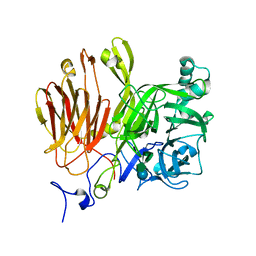 | |
3LDR
 
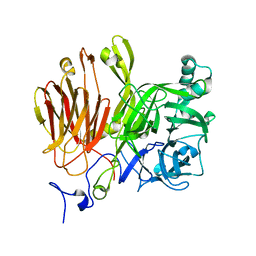 | |
3BH4
 
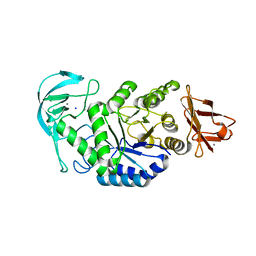 | | High resolution crystal structure of Bacillus amyloliquefaciens alpha-amylase | | Descriptor: | Alpha-amylase, CALCIUM ION, SODIUM ION | | Authors: | Alikhajeh, J, Khajeh, K, Ranjbar, B, Naderi-Manesh, H, Lin, Y.H, Liu, M.Y, Chen, C.J. | | Deposit date: | 2007-11-28 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Bacillus amyloliquefaciens alpha-amylase at high resolution: implications for thermal stability.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5XGT
 
 | |
5XVS
 
 | | Crystal structure of UDP-GlcNAc 2-epimerase NeuC complexed with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), LITHIUM ION, ... | | Authors: | Ko, T.P, Hsieh, T.J, Chen, S.C, Wu, S.C, Guan, H.H, Yang, C.H, Chen, C.J, Chen, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
4OKW
 
 | | Crystal structure of W741L-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, R-BICALUTAMIDE, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-23 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OEA
 
 | | Crystal structure of AR-LBD | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-12 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OFR
 
 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-15 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OFU
 
 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-15 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OK1
 
 | | Crystal structure of W741L-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, R-BICALUTAMIDE, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-21 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OIU
 
 | | Crystal structure of T877A-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, HYDROXYFLUTAMIDE, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-20 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OKT
 
 | | Crystal structure of W741L-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, R-BICALUTAMIDE, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-22 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OLM
 
 | | Crystal structure of W741L-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, R-BICALUTAMIDE, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-24 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OJB
 
 | | Crystal structure of W741L-AR-LBD | | Descriptor: | Androgen receptor, R-BICALUTAMIDE | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-21 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OED
 
 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Protein BUD31 homolog, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
