1Y0Y
 
 | |
1Y0R
 
 | |
7NC1
 
 | |
7NCB
 
 | | Glutathione-S-transferase GliG mutant H26A | | Descriptor: | Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCD
 
 | | Glutathione-S-transferase GliG mutant N27D | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glutathione S-transferase GliG, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC9
 
 | | Glutathione-S-transferase GliG mutant H26N | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCM
 
 | | Glutathione-S-transferase GliG mutant E82A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCU
 
 | |
7NCT
 
 | | Glutathione-S-transferase GliG mutant K127G | | Descriptor: | Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC8
 
 | | Glutathione-S-transferase GliG mutant S24A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC6
 
 | | Glutathione-S-transferase GliG in complex with cyclo[L-Phe-L-Ser]-bis-glutathione-adduct | | Descriptor: | (2~{S})-2-azanyl-5-[[(2~{R})-3-[(2~{R},5~{R})-5-(hydroxymethyl)-3,6-bis(oxidanylidene)-2-(phenylmethyl)-5-sulfanyl-piperazin-2-yl]sulfanyl-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCO
 
 | | Glutathione-S-transferase GliG mutant K127R | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC2
 
 | |
7NCE
 
 | | Glutathione-S-transferase GliG mutant N27A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCL
 
 | | Glutathione-S-transferase GliG mutant E82Q | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC5
 
 | |
7NCN
 
 | | Glutathione-S-transferase GliG mutant S83A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC3
 
 | |
7NCP
 
 | | Glutathione-S-transferase GliG mutant K127A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG, SODIUM ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5DAV
 
 | |
5NRU
 
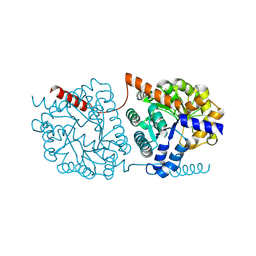 | | Cys-Gly dipeptidase GliJ in complex with Zn2+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, ZINC ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NS5
 
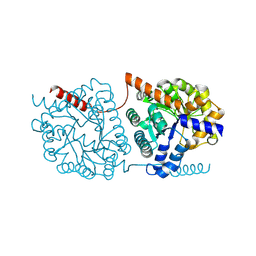 | | Cys-Gly dipeptidase GliJ in complex with Cu2+ and Zn2+ | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Dipeptidase gliJ, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NS1
 
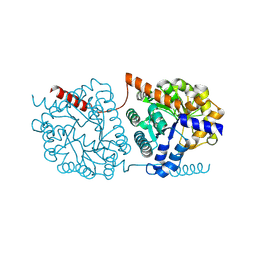 | | Cys-Gly dipeptidase GliJ in complex with Ni2+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, NICKEL (II) ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NRZ
 
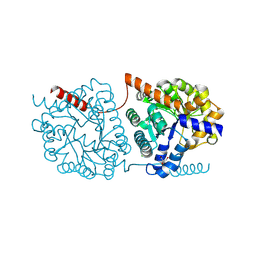 | | Cys-Gly dipeptidase GliJ in complex with Mn2+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, GLYCEROL, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NRT
 
 | | Cys-Gly dipeptidase GliJ in complex with Ca2+ | | Descriptor: | CALCIUM ION, Dipeptidase gliJ, MAGNESIUM ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
