3WDZ
 
 | | Crystal Structure of Keap1 in Complex with phosphorylated p62 | | Descriptor: | Kelch-like ECH-associated protein 1, Peptide from Sequestosome-1 | | Authors: | Fukutomi, T, Takagi, K, Mizushima, T, Tanaka, K, Komatsu, M, Yamamoto, M. | | Deposit date: | 2013-06-26 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy.
Mol.Cell, 51, 2013
|
|
3B0F
 
 | | Crystal structure of the UBA domain of p62 and its interaction with ubiquitin | | Descriptor: | SULFATE ION, Sequestosome-1 | | Authors: | Isogai, S, Morimoto, D, Arita, K, Unzai, S, Tenno, T, Hasegawa, J, Sou, Y, Komatsu, M, Tanaka, K, Shirakawa, M, Tochio, H. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2D1I
 
 | | Structure of human Atg4b | | Descriptor: | Cysteine protease APG4B | | Authors: | Kumanomidou, T, Mizushima, T, Komatsu, M, Suzuki, A, Tanida, I, Sou, Y.S, Ueno, T, Kominami, E, Tanaka, K, Yamane, T. | | Deposit date: | 2005-08-24 | | Release date: | 2006-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Atg4b, a Processing and De-conjugating Enzyme for Autophagosome-forming Modifiers
J.Mol.Biol., 355, 2006
|
|
2E33
 
 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, Ribonuclease pancreatic, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E31
 
 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, S-phase kinase-associated protein 1A | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E32
 
 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, S-phase kinase-associated protein 1A | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Z84
 
 | | Insights from crystal and solution structures of mouse UfSP1 | | Descriptor: | Ufm1-specific protease 1 | | Authors: | Ha, B.H, Ahn, H.C, Kang, S.H, Tanaka, K, Chung, C.H, Kim, E.E. | | Deposit date: | 2007-08-30 | | Release date: | 2008-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for Ufm1 processing by UfSP1
J. Biol. Chem., 283, 2008
|
|
2ZJD
 
 | | Crystal Structure of LC3-p62 complex | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B precursor, undecameric peptide from Sequestosome-1 | | Authors: | Ichimura, Y, Kumanomidou, T, Sou, Y, Mizushima, T, Ezaki, J, Ueno, T, Kominami, E, Yamane, T, Tanaka, K, Komatsu, M. | | Deposit date: | 2008-03-05 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Basis for Sorting Mechanism of p62 in Selective Autophagy
J.Biol.Chem., 283, 2008
|
|
3ASE
 
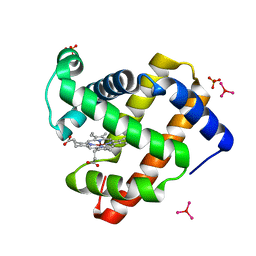 | | Crystal Structure of Zinc myoglobin soaked with Ru3O cluster | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING ZN, SULFATE ION, ... | | Authors: | Koshiyama, T, Shirai, M, Hikage, T, Tabe, H, Tanaka, K, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-12-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Post-Crystal Engineering of Zinc-Substituted Myoglobin to Construct a Long-Lived Photoinduced Charge-Separation System
Angew.Chem.Int.Ed.Engl., 2011
|
|
5GWF
 
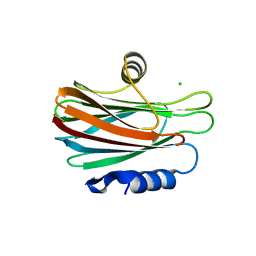 | | FraC with GlcNAc(6S) bound | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, CHLORIDE ION, DELTA-actitoxin-Afr1a, ... | | Authors: | Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2016-09-11 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Haemolytic actinoporins interact with carbohydrates using their lipid-binding module
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 372, 2017
|
|
4WDC
 
 | |
7V1W
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium in complex with beta-D-arabinofuranose | | Descriptor: | CALCIUM ION, Difructose dianhydride I synthase/hydrolase (alphaFFase1), beta-D-arabinofuranose | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
7V1V
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium, ligand-free form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
7V1X
 
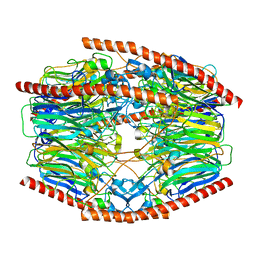 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium in complex with beta-D-fructofuranose | | Descriptor: | CALCIUM ION, Difructose dianhydride I synthase/hydrolase, beta-D-fructofuranose | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
4N4G
 
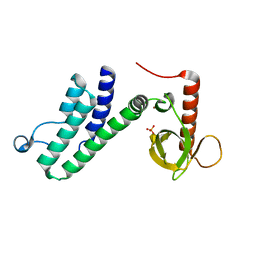 | |
4N4I
 
 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.3K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.3, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
4N4H
 
 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.1K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.1, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
6YCQ
 
 | | Crystal structure of the DNA binding domain of Arabidopsis thaliana Auxin Response Factor 1 (AtARF1) in complex with High Affinity DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 21-7A, 21-7B, ... | | Authors: | Crespo, I, Weijers, D, Boer, D.R. | | Deposit date: | 2020-03-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8HHV
 
 | | endo-alpha-D-arabinanase EndoMA1 from Microbacterium arabinogalactanolyticum | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Nakashima, C, Li, J, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2022-11-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
3L2D
 
 | |
3L2E
 
 | | Glycocyamine kinase, alpha-beta heterodimer from marine worm Namalycastis sp. | | Descriptor: | Glycocyamine kinase alpha chain, Glycocyamine kinase beta chain | | Authors: | Lim, K, Pullalarevu, S, Herzberg, O. | | Deposit date: | 2009-12-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
Biochemistry, 49, 2010
|
|
7YNE
 
 | |
4V7N
 
 | | Glycocyamine kinase, beta-beta homodimer from marine worm Namalycastis sp., with transition state analog Mg(II)-ADP-NO3-glycocyamine. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANIDINO ACETATE, Glycocyamine kinase beta chain, ... | | Authors: | Lim, K, Pullalarevu, S, Herzberg, O. | | Deposit date: | 2009-12-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
Biochemistry, 49, 2010
|
|
3OQC
 
 | |
8IC1
 
 | | endo-alpha-D-arabinanase EndoMA1 D51N mutant from Microbacterium arabinogalactanolyticum in complex with arabinooligosaccharides | | Descriptor: | (3~{a}~{S},5~{R},6~{R},6~{a}~{S})-5-(hydroxymethyl)-2,2-dimethyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]dioxol-6-ol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, J, Nakashima, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
