7BIS
 
 | |
7BK9
 
 | |
2XUV
 
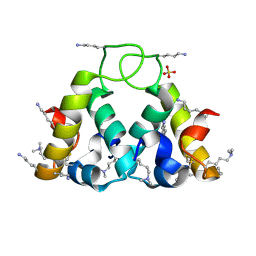 | | The structure of HdeB | | Descriptor: | HDEB, SULFATE ION | | Authors: | Naismith, J.H, Wang, W. | | Deposit date: | 2010-10-21 | | Release date: | 2011-08-24 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Salt Bridges Regulate Both Dimer Formation and Monomeric Flexibility in Hdeb and May Have a Role in Periplasmic Chaperone Function.
J.Mol.Biol., 415, 2012
|
|
6SJ1
 
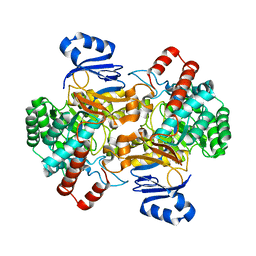 | | Amidohydrolase, AHS | | Descriptor: | Amidohydrolase, ZINC ION | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SIW
 
 | | PaaK family AMP-ligase with AMP | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SSG
 
 | | Transaminase with DCS bound | | Descriptor: | ForI-DCS, SULFATE ION, [4-[(~{Z})-[(2~{R},5~{R})-5-(azanyloxymethyl)-3,6-bis(oxidanylidene)piperazin-2-yl]methoxyiminomethyl]-6-methyl-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2019-09-06 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PMP-diketopiperazine adducts form at the active site of a PLP dependent enzyme involved in formycin biosynthesis.
Chem.Commun.(Camb.), 55, 2019
|
|
6SSE
 
 | | Transaminase with PMP bound | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ForI-PMP, SULFATE ION | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2019-09-06 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | PMP-diketopiperazine adducts form at the active site of a PLP dependent enzyme involved in formycin biosynthesis.
Chem.Commun.(Camb.), 55, 2019
|
|
6SIY
 
 | | PaaK family AMP-ligase with AMP and substrate | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYANTHRANILIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SJ0
 
 | | Amidohydrolase, AHS | | Descriptor: | Amidohydrolase, BICARBONATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SSF
 
 | | Transaminase with LCS bound | | Descriptor: | ForI-LCS, SULFATE ION, [4-[(~{Z})-[(2~{S},5~{S})-5-(azanyloxymethyl)-3,6-bis(oxidanylidene)piperazin-2-yl]methoxyiminomethyl]-6-methyl-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2019-09-06 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PMP-diketopiperazine adducts form at the active site of a PLP dependent enzyme involved in formycin biosynthesis.
Chem.Commun.(Camb.), 55, 2019
|
|
6SSD
 
 | | Transaminase with PLP bound | | Descriptor: | ForI-PLP, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2019-09-06 | | Release date: | 2020-01-15 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | PMP-diketopiperazine adducts form at the active site of a PLP dependent enzyme involved in formycin biosynthesis.
Chem.Commun.(Camb.), 55, 2019
|
|
6TM4
 
 | | NatL2 in complex with two molecules of salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SIZ
 
 | | PaaK family AMP-ligase with ANP and substrate | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYANTHRANILIC ACID, AMP-dependent synthetase and ligase, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SJ3
 
 | | Amidohydrolase, AHS with 3-HBA | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYBENZOIC ACID, Amidohydrolase, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SIX
 
 | | PaaK family AMP-ligase with ANP | | Descriptor: | 1,2-ETHANEDIOL, AMP-dependent synthetase and ligase, CITRATE ANION, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SJ2
 
 | | Amidohydrolase, AHS with 3-HAA | | Descriptor: | 3-HYDROXYANTHRANILIC ACID, Amidohydrolase, GLYCEROL, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1FQ0
 
 | | KDPG ALDOLASE FROM ESCHERICHIA COLI | | Descriptor: | CITRIC ACID, KDPG ALDOLASE | | Authors: | Naismith, J.H. | | Deposit date: | 2000-09-01 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Directed evolution of a new catalytic site in 2-keto-3-deoxy-6-phosphogluconate aldolase from Escherichia coli.
Structure, 9, 2001
|
|
1FWR
 
 | |
1QGL
 
 | |
2VG1
 
 | | Rv1086 E,E-farnesyl diphosphate complex | | Descriptor: | FARNESYL DIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2VL7
 
 | | Structure of S. tokodaii Xpd4 | | Descriptor: | PHOSPHATE ION, XPD | | Authors: | Naismith, J.H, Johnson, K.A, Oke, M, McMahon, S.A, Liu, L, White, M.F, Zawadski, M, Carter, L.G. | | Deposit date: | 2008-01-08 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the DNA Repair Helicase Xpd.
Cell(Cambridge,Mass.), 133, 2008
|
|
2V7I
 
 | | PrnB native | | Descriptor: | PRNB, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Naismith, J.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway is Related to the Heme-Dependent Dioxygenase Superfamily
Biochemistry, 46, 2007
|
|
2VFW
 
 | | Rv1086 native | | Descriptor: | SHORT-CHAIN Z-ISOPRENYL DIPHOSPHATE SYNTHETASE, SULFATE ION | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-05 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2V81
 
 | | Native KDPGal structure | | Descriptor: | 2-DEHYDRO-3-DEOXY-6-PHOSPHOGALACTONATE ALDOLASE | | Authors: | Naismith, J.H. | | Deposit date: | 2007-08-02 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization and crystal structure of Escherichia coli KDPGal aldolase.
Bioorg. Med. Chem., 16, 2008
|
|
2VG2
 
 | | Rv2361 with IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, DIPHOSPHATE, ... | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
