3FP4
 
 | | Crystal structure of Tom71 complexed with Ssa1 C-terminal fragment | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
3T0H
 
 | |
3T0Z
 
 | | Hsp90 N-terminal domain bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Li, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90.
Acta Biochim Biophys Sin (Shanghai), 44, 2012
|
|
3T10
 
 | | HSP90 N-terminal domain bound to ACP | | Descriptor: | Heat shock protein HSP 90-alpha, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Li, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90.
Acta Biochim Biophys Sin (Shanghai), 44, 2012
|
|
7WJS
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13157 | | Descriptor: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WKY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13153 | | Descriptor: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-12 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WNI
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13158 | | Descriptor: | 1,2-ETHANEDIOL, 7-[2-[2,4-bis(fluoranyl)phenoxy]-5-(2-oxidanylpropan-2-yl)phenyl]-2-[4-(2-hydroxyethyloxy)-3,5-dimethyl-phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WMU
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13146 | | Descriptor: | 1,2-ETHANEDIOL, Isoform 4 of Bromodomain-containing protein 2, ~{N}-[4-[2,4-bis(fluoranyl)phenoxy]-3-[2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-5-methyl-4-oxidanylidene-furo[3,2-c]pyridin-7-yl]phenyl]ethanesulfonamide | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WN5
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13142 | | Descriptor: | 1,2-ETHANEDIOL, Isoform 4 of Bromodomain-containing protein 2, POTASSIUM ION, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WLN
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13153 | | Descriptor: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WNA
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13120 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Isoform 4 of Bromodomain-containing protein 2, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WMQ
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13157 | | Descriptor: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-16 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
1C8U
 
 | |
8WY4
 
 | | GajA tetramer with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X5I
 
 | | tetramer Gabija with ATP (local refinement) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, MAGNESIUM ION | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X5N
 
 | | Tetramer Gabija with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, Gabija protein GajB, ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X51
 
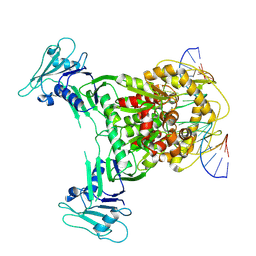 | | Cryo-EM structure of Gabija GajA in complex with DNA(focused refinement) | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
3RY7
 
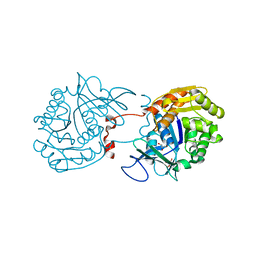 | | Crystal Structure of Sa239 | | Descriptor: | GLYCEROL, Ribokinase | | Authors: | Li, J, Wu, M, Wang, L, Zang, J. | | Deposit date: | 2011-05-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Sa239 reveals the structural basis for the activation of ribokinase by monovalent cations.
J.Struct.Biol., 177, 2012
|
|
6CYZ
 
 | |
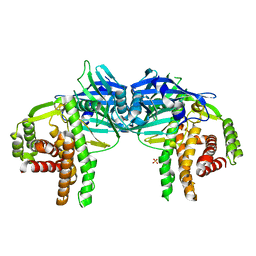
4O7P
 
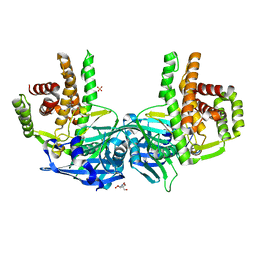 | | Crystal structure of Mycobacterium tuberculosis maltose kinase MaK complexed with maltose | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Maltokinase, SULFATE ION, ... | | Authors: | Li, J, Guan, X.T, Rao, Z.H. | | Deposit date: | 2013-12-26 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Homotypic dimerization of a maltose kinase for molecular scaffolding.
Sci Rep, 4, 2014
|
|
4O7O
 
 | |
8EW5
 
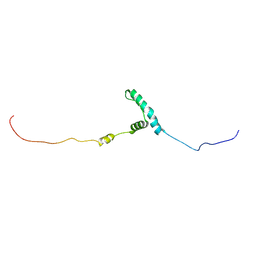 | | The structure of flightin within myosin thick filaments from Bombus ignitus flight muscle | | Descriptor: | Flightin | | Authors: | Li, J, Rahmani, H, Abbasi Yeganeh, F, Rastegarpouyani, H, Taylor, D.W, Wood, N.B, Previs, M.J, Iwamoto, H, Taylor, K.A. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-04 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of the Flight Muscle Thick Filament from the Bumble Bee, Bombus ignitus , at 6 angstrom Resolution.
Int J Mol Sci, 24, 2022
|
|
7WNT
 
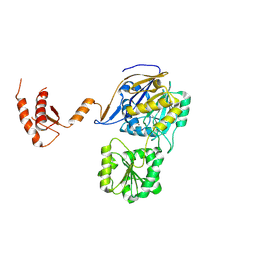 | | RNase J from Mycobacterium tuberculosis | | Descriptor: | Ribonuclease J, ZINC ION | | Authors: | Li, J, Bao, L, Hu, J, Zhan, B, Li, Z. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural ans functional study of RNase J from Mycobacterium tuberculosis
To Be Published
|
|
7WNU
 
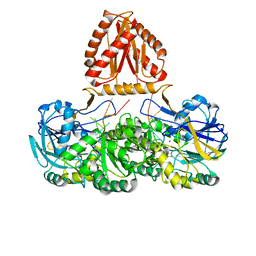 | | Mycobacterium tuberculosis Rnase J complex with 7nt RNA | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*AP*A)-3'), Ribonuclease J, ZINC ION | | Authors: | Li, J, Hu, J, Bao, L, Zhan, B, Li, Z. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural ans functional study of RNase J from Mycobacterium tuberculosis
To Be Published
|
|
2AD6
 
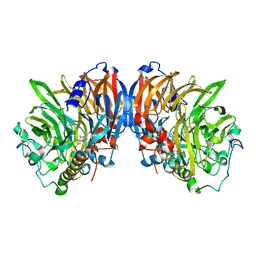 | | crystal structure of methanol dehydrogenase from M. W3A1 (form C) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase subunit 1, Methanol dehydrogenase subunit 2, ... | | Authors: | Li, J, Gan, J.-H, Xia, Z.-X, Mathews, F.S. | | Deposit date: | 2005-07-20 | | Release date: | 2006-07-25 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase
Biochem.Biophys.Res.Commun., 406, 2011
|
|
