3NLY
 
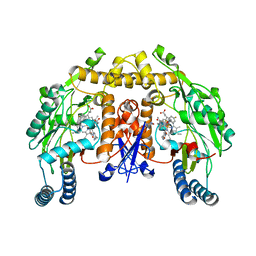 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2- (2,2-Difluoro-2-(4-fluorophenyl)ethylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[2,2-difluoro-2-(4-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
3NLZ
 
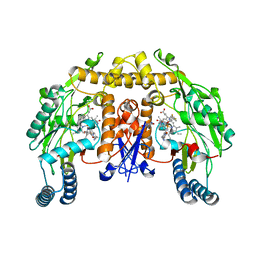 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2-(2,2-Difluoro-2-phenylethylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-{2-[(2,2-difluoro-2-phenylethyl)amino]ethoxy}pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
5O77
 
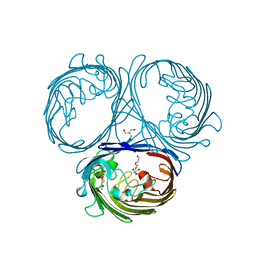 | | Klebsiella pneumoniae OmpK35 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, OmpK35 | | Authors: | van den berg, B, Pathania, M, Zahn, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
3NLX
 
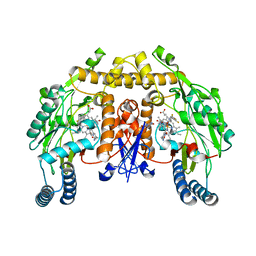 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2- (2,2-Difluoro-2-(3-fluorophenyl)ethylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[2,2-difluoro-2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
1O1Z
 
 | |
1O2D
 
 | |
1O4W
 
 | |
1O4U
 
 | |
1O1X
 
 | |
1O1Y
 
 | |
3NM0
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2-(2,2-Difluoro-2-phenylethylamino)ethoxy) pyrrolidin-3-yl)methyl)-4-methyl-3,4,5,6-tetrahydropyridin-2-amine | | Descriptor: | (4S,6S)-6-{[(3R,4R)-4-{2-[(2,2-difluoro-2-phenylethyl)amino]ethoxy}pyrrolidin-3-yl]methyl}-4-methyl-3,4,5,6-tetrahydropyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
3FAH
 
 | | Glycerol inhibited form of Aldehyde oxidoreductase from Desulfovibrio gigas | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), Aldehyde oxidoreductase, CHLORIDE ION, ... | | Authors: | Santos-Silva, T, Romao, M.J. | | Deposit date: | 2008-11-17 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetic, structural, and EPR studies reveal that aldehyde oxidoreductase from Desulfovibrio gigas does not need a sulfido ligand for catalysis and give evidence for a direct Mo-C interaction in a biological system.
J.Am.Chem.Soc., 131, 2009
|
|
3FC4
 
 | | Ethylene glycol inhibited form of Aldehyde oxidoreductase from Desulfovibrio gigas | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), 1,2-ETHANEDIOL, Aldehyde oxidoreductase, ... | | Authors: | Santos-Silva, T, Romao, M.J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Kinetic, structural, and EPR studies reveal that aldehyde oxidoreductase from Desulfovibrio gigas does not need a sulfido ligand for catalysis and give evidence for a direct Mo-C interaction in a biological system.
J.Am.Chem.Soc., 131, 2009
|
|
2E6W
 
 | | Solution structure and calcium binding properties of EF-hands 3 and 4 of calsenilin | | Descriptor: | CALCIUM ION, Calsenilin | | Authors: | Yu, L, Sun, C, Mendoza, R, Hebert, E, Pereda-Lopez, A, Hajduk, P.J, Olejniczak, E.T. | | Deposit date: | 2007-01-05 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and calcium-binding properties of EF-hands 3 and 4 of calsenilin.
Protein Sci., 16, 2007
|
|
7XJT
 
 | |
7X99
 
 | |
2PS7
 
 | | Y295F trichodiene synthase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Trichodiene synthase | | Authors: | Vedula, L.S, Cane, D.E, Christianson, D.W. | | Deposit date: | 2007-05-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and mechanistic analysis of trichodiene synthase using site-directed mutagenesis: probing the catalytic function of tyrosine-295 and the asparagine-225/serine-229/glutamate-233-Mg2+B motif.
Arch.Biochem.Biophys., 469, 2008
|
|
3NNY
 
 | | Structure of rat neuronal nitric oxide synthase heme domain complexed with 6-(((3R,4R)-4-(2-(3-Fluorophenethylamino)ethoxy)pyrrolidin-3-yl)methyl)pyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}pyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2010-06-24 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peripheral but crucial: a hydrophobic pocket (Tyr(706), Leu(337), and Met(336)) for potent and selective inhibition of neuronal nitric oxide synthase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3B3O
 
 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-AMINO-4-METHYLPYRIDIN-2-YL)METHYL]PYRROLIDIN-3-YL}-N'-(4-CHLOROBENZYL)ETHANE-1,2-DIAMINE, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
1O3U
 
 | |
1O22
 
 | |
5O79
 
 | | Klebsiella pneumoniae OmpK36 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, OmpK36 | | Authors: | van den berg, B, Pathania, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
1O4V
 
 | |
3BVM
 
 | |
3BYY
 
 | |
