6PT4
 
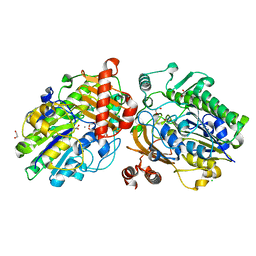 | | Crystal structure of apo PsS1_NC | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2019-07-14 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the kappa / iota-carrageenan metabolism pathway of some marinePseudoalteromonasspecies.
Commun Biol, 2, 2019
|
|
6PZZ
 
 | | CryoEM derived model of NA-80 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-80 fragment antibody heavy chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6Q1P
 
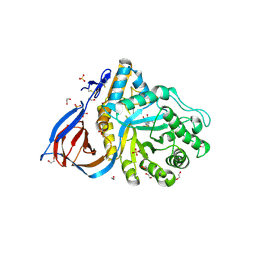 | | Glucocerebrosidase in complex with pharmacological chaperone norIMX8 | | Descriptor: | (2S,3S,4S,5R)-2-{[(4-methylpentyl)sulfonyl]methyl}piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vickers, C, Withers, S.G, Boraston, A.B. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Pharmacological chaperones for GCase that switch conformation with pH enhance enzyme levels in Gaucher animal models.
To Be Published
|
|
1G6O
 
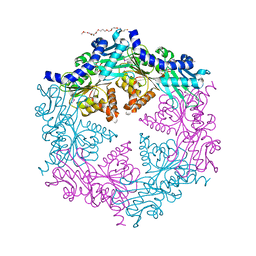 | | CRYSTAL STRUCTURE OF THE HELICOBACTER PYLORI ATPASE, HP0525, IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CAG-ALPHA, DI(HYDROXYETHYL)ETHER | | Authors: | Yeo, H.J, Savvides, S.N, Herr, A.B, Lanka, E, Waksman, G, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-11-07 | | Release date: | 2001-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system.
Mol.Cell, 6, 2000
|
|
6QKD
 
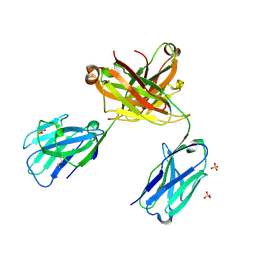 | | CRYSTAL STRUCTURE OF vhh-based FAB-fragment of antibody BCD-085 | | Descriptor: | CHLORIDE ION, FAB HEAVY CHAIN, FAB LIGHT CHAIN, ... | | Authors: | Kostareva, O.S, Kolyadenko, I.A, Ulitin, A.B, Ekimova, V.M, Evdokimov, S.R, Garber, M.B, Tishchenko, T.V, Gabdulkhakov, A.G. | | Deposit date: | 2019-01-29 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fab Fragment of VHH-Based Antibody Netakimab: Crystal Structure and Modeling Interaction with Cytokine IL-17A
Crystals, 2019
|
|
6Q44
 
 | | Est3 telomerase subunit in the yeast Hansenula polymorpha | | Descriptor: | Uncharacterized protein | | Authors: | Mantsyzov, A.B, Mariasina, S.S, Petrova, O.A, Efimov, S.V, Dontsova, O.A, Polshakov, V.I. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-25 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Insights into the structure and function of Est3 from the Hansenula polymorpha telomerase.
Sci Rep, 10, 2020
|
|
1GP3
 
 | | Human IGF2R domain 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Brown, J, Esnouf, R.M, Jones, M.A, Linnell, J, Harlos, K, Hassan, A.B, Jones, E.Y. | | Deposit date: | 2001-10-29 | | Release date: | 2002-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a Functional Igf2R Fragment Determined from the Anomalous Scattering of Sulfur
Embo J., 21, 2002
|
|
6R2W
 
 | | Crystal structure of the super-active FVIIa variant VYT in complex with tissue factor | | Descriptor: | CALCIUM ION, Coagulation factor VII, N-acetyl-D-phenylalanyl-N-[(2S,3S)-6-carbamimidamido-1-chloro-2-hydroxyhexan-3-yl]-L-phenylalaninamide, ... | | Authors: | Sorensen, A.B, Svensson, L.A, Gandhi, P.S. | | Deposit date: | 2019-03-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Beating tissue factor at its own game: Design and properties of a soluble tissue factor-independent coagulation factor VIIa.
J.Biol.Chem., 295, 2020
|
|
1GP0
 
 | | Human IGF2R domain 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, SULFATE ION | | Authors: | Brown, J, Esnouf, R.M, Jones, M.A, Linnell, J, Harlos, K, Hassan, A.B, Jones, E.Y. | | Deposit date: | 2001-10-29 | | Release date: | 2002-02-28 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a Functional Igf2R Fragment Determined from the Anomalous Scattering of Sulfur
Embo J., 21, 2002
|
|
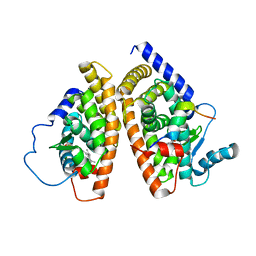
1HR9
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Malate Dehydrogenase Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MALATE DEHYDROGENASE, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
6RBB
 
 | | CRYSTAL STRUCTURE OF the VhH-domain of anti-IL-17A antibody netakimab | | Descriptor: | VHH domain of netakimab | | Authors: | Kostareva, O.S, Kolyadenko, I.A, Ulitin, A.B, Ekimova, V.M, Evdokimov, S.R, Garber, M.B, Tishchenko, T.V, Gabdulkhakov, A.G. | | Deposit date: | 2019-04-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | CRYSTAL STRUCTURE OF the VhH-domain of anti-IL-17A antibody netakimab
To Be Published
|
|
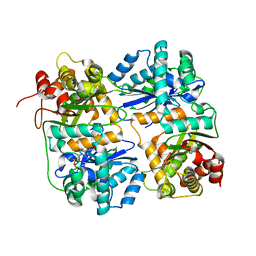
1FM6
 
 | | THE 2.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE HETERODIMER OF THE HUMAN RXRALPHA AND PPARGAMMA LIGAND BINDING DOMAINS RESPECTIVELY BOUND WITH 9-CIS RETINOIC ACID AND ROSIGLITAZONE AND CO-ACTIVATOR PEPTIDES. | | Descriptor: | (9cis)-retinoic acid, 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), ... | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Miller, A.B, Bledsoe, R.K, Milburn, M.V, Kliewer, S.A, Willson, T.M, Xu, H.E. | | Deposit date: | 2000-08-16 | | Release date: | 2001-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors.
Mol.Cell, 5, 2000
|
|
1HR6
 
 | | Yeast Mitochondrial Processing Peptidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
6PV4
 
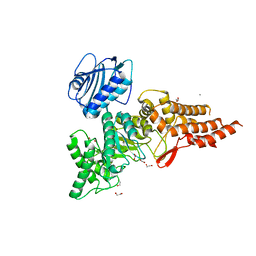 | | Structure of CpGH84A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycoside Hydrolase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of four family 84 glycoside hydrolases from the opportunistic pathogen Clostridium perfringens.
Glycobiology, 30, 2019
|
|
1HR8
 
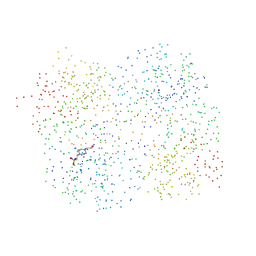 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Cytochrome C Oxidase IV Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYTOCHROME C OXIDASE POLYPEPTIDE IV, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
6PZY
 
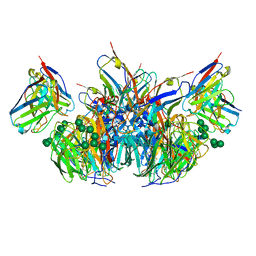 | | CryoEM derived model of NA-73 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-73 fragment antibody heavy chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6Q1N
 
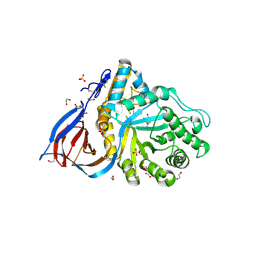 | | Glucocerebrosidase in complex with pharmacological chaperone IMX8 | | Descriptor: | (2R,3S,4S,5R)-2-[2-(methylsulfanyl)ethyl]piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vickers, C, Withers, S.G, Boraston, A.B. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2.526 Å) | | Cite: | Pharmacological chaperones for GCase that switch conformation with pH enhance enzyme levels in Gaucher animal models.
To Be Published
|
|
1HZS
 
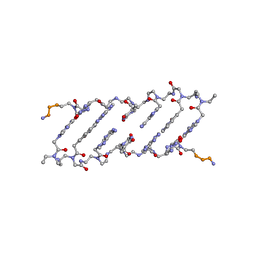 | | Crystal structure of a peptide nucleic acid duplex (BT-PNA) containing a bicyclic analogue of thymine | | Descriptor: | PEPTIDE NUCLEIC ACID | | Authors: | Eldrup, A.B, Nielsen, B.B, Haaima, G, Rasmussen, H, Kastrup, J.S, Christensen, C, Nielsen, P.E. | | Deposit date: | 2001-01-26 | | Release date: | 2001-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 1,8-Naphthyridin-2(1H)-ones. Novel Bicyclic and Tricyclic Analogues of Thymine in Peptide Nucleic Acids (PNAs)
Eur.J.Org.Chem., 9, 2001
|
|
6PTM
 
 | |
1FM9
 
 | | THE 2.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE HETERODIMER OF THE HUMAN RXRALPHA AND PPARGAMMA LIGAND BINDING DOMAINS RESPECTIVELY BOUND WITH 9-CIS RETINOIC ACID AND GI262570 AND CO-ACTIVATOR PEPTIDES. | | Descriptor: | (9cis)-retinoic acid, 2-(2-BENZOYL-PHENYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA, ... | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Miller, A.B, Bledsoe, R.K, Milburn, M.V, Kliewer, S.A, Willson, T.M, Xu, H.E. | | Deposit date: | 2000-08-16 | | Release date: | 2001-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors.
Mol.Cell, 5, 2000
|
|
1I1G
 
 | | CRYSTAL STRUCTURE OF THE LRP-LIKE TRANSCRIPTIONAL REGULATOR FROM THE ARCHAEON PYROCOCCUS FURIOSUS | | Descriptor: | TRANSCRIPTIONAL REGULATOR LRPA | | Authors: | Leonard, P.M, Smits, S.H.J, Sedelnikova, S.E, Brinkman, A.B, de Vos, W.M, van der Oost, J, Rice, D.W, Rafferty, J.B. | | Deposit date: | 2001-02-01 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus.
EMBO J., 20, 2001
|
|
1IBY
 
 | | RED COPPER PROTEIN NITROSOCYANIN FROM NITROSOMONAS EUROPAEA | | Descriptor: | COPPER (II) ION, HEXANE-1,6-DIOL, NITROSOCYANIN | | Authors: | Lieberman, R.L, Arciero, D.M, Hooper, A.B, Rosenzweig, A.C. | | Deposit date: | 2001-03-29 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a novel red copper protein from Nitrosomonas europaea.
Biochemistry, 40, 2001
|
|
6PSO
 
 | | Crystal structure of PsS1_19B C77S in complex with iota-neocarratetraose | | Descriptor: | 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2019-07-13 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the kappa / iota-carrageenan metabolism pathway of some marinePseudoalteromonasspecies.
Commun Biol, 2, 2019
|
|
6PT6
 
 | | Crystal structure of PsS1_NC C84S in complex with i-neocarratetraose | | Descriptor: | 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, exo-2S-iota carrageenan S1 sulfatase | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2019-07-15 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into the kappa / iota-carrageenan metabolism pathway of some marinePseudoalteromonasspecies.
Commun Biol, 2, 2019
|
|
6PZ8
 
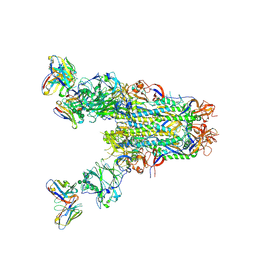 | | MERS S0 trimer in complex with variable domain of antibody G2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G2 heavy chain, ... | | Authors: | Bowman, C.A, Pallesen, J, Ward, A.B. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD.
Cell Rep, 28, 2019
|
|
