2L42
 
 | |
2LI6
 
 | |
6IZG
 
 | |
5GO0
 
 | |
2L7Y
 
 | |
2LJ8
 
 | |
2M1H
 
 | | Solution structure of a PWWP domain from Trypanosoma brucei | | Descriptor: | Transcription elongation factor S-II | | Authors: | Wang, R, Fan, K, Liao, S, Zhang, J, Tu, X. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of TbTFIIS2-1 PWWP domain from Trypanosoma brucei.
Proteins, 84, 2016
|
|
2NAS
 
 | |
2L32
 
 | |
2LRW
 
 | | Solution structure of a ubiquitin-like protein from Trypanosoma brucei | | Descriptor: | Ubiquitin, putative | | Authors: | Wang, R, Wang, T, Liao, S, Zhang, J, Tu, X. | | Deposit date: | 2012-04-13 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a ubiqutin-like protein from Trypanosoma bucei
To be Published
|
|
6IF4
 
 | | Crystal structure of Tbtudor | | Descriptor: | Histone acetyltransferase | | Authors: | Gao, J, Ye, K, Diwu, Y, Liao, S, Tu, X. | | Deposit date: | 2018-09-18 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.934 Å) | | Cite: | Crystal structure of TbEsa1 presumed Tudor domain from Trypanosoma brucei.
J.Struct.Biol., 209, 2020
|
|
7FAW
 
 | | Structure of LW domain from Yeast | | Descriptor: | Transcription elongation factor S-II | | Authors: | Liao, S, Gao, J, Tu, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for evolutionarily conserved interactions between TFIIS and Paf1C.
Int.J.Biol.Macromol., 253, 2023
|
|
7FAX
 
 | |
7DL8
 
 | |
6LEN
 
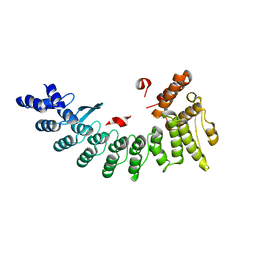 | | Structure of NS11 bound FEM1C | | Descriptor: | Protein fem-1 homolog C,NS11 peptide | | Authors: | Chen, x, Liao, S, Xu, C. | | Deposit date: | 2019-11-25 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LF0
 
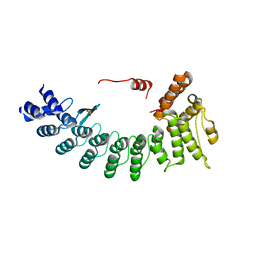 | | Structure of FEM1C | | Descriptor: | Protein fem-1 homolog C, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LBN
 
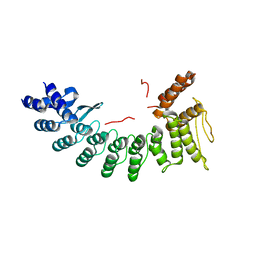 | | Structure of SIL1-bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Nucleotide exchange factor SIL1 | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LEY
 
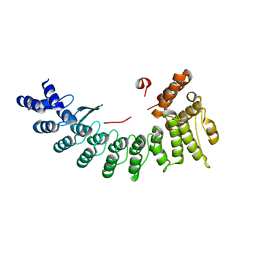 | | Structure of Sil1G bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Nucleotide exchange factor SIL1 | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
3MTS
 
 | | Chromo Domain of Human Histone-Lysine N-Methyltransferase SUV39H1 | | Descriptor: | Histone-lysine N-methyltransferase SUV39H1 | | Authors: | Lam, R, Li, Z, Wang, J, Crombet, L, Walker, J.R, Ouyang, H, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human SUV39H1 Chromodomain and Its Recognition of Histone H3K9me2/3.
Plos One, 7, 2012
|
|
1VB0
 
 | | Atomic resolution structure of atratoxin-b, one short-chain neurotoxin from Naja atra | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cobrotoxin b, SULFATE ION | | Authors: | Lou, X, Liu, Q, Teng, M, Niu, L, Huang, Q, Hao, Q. | | Deposit date: | 2004-02-20 | | Release date: | 2004-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | The atomic resolution crystal structure of atratoxin determined by single wavelength anomalous diffraction phasing
J.Biol.Chem., 279, 2004
|
|
7YGG
 
 | |
3JYT
 
 | | K65R mutant HIV-1 reverse transcriptase cross-linked to DS-DNA and complexed with DATP as the incoming nucleotide substrate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(*A*TP*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2009-09-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the role of the K65r mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance.
J.Biol.Chem., 284, 2009
|
|
3JSM
 
 | | K65R mutant HIV-1 reverse transcriptase cross-linked to DS-DNA and complexed with tenofovir-diphosphate as the incoming nucleotide substrate | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(*A*TP*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), HIV-1 REVERSE TRANSCRIPTASE P51 SUBUNIT, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2009-09-10 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the role of the K65r mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance.
J.Biol.Chem., 284, 2009
|
|
4R5P
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and a nucleoside triphosphate mimic alpha-carboxy nucleoside phosphonate inhibitor | | Descriptor: | 5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3', 5'-D(*TP*GP*GP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3', HIV-1 reverse transcriptase, ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-08-21 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Alpha-carboxy nucleoside phosphonates as universal nucleoside triphosphate mimics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5X5S
 
 | |
