6R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441D MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
5R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441A MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
7R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441Q MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
1R1R
 
 | |
1HK8
 
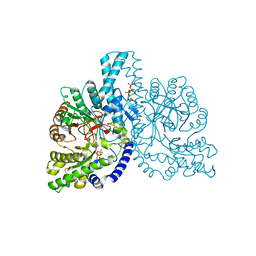 | | STRUCTURAL BASIS FOR ALLOSTERIC SUBSTRATE SPECIFICITY REGULATION IN CLASS III RIBONUCLEOTIDE REDUCTASES: NRDD IN COMPLEX WITH DGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOTIDE-TRIPHOSPHATE REDUCTASE, MANGANESE (II) ION, ... | | Authors: | Larsson, K.-M, Andersson, J, Sjoeberg, B.-M, Nordlund, P, Logan, D.T. | | Deposit date: | 2003-03-06 | | Release date: | 2003-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Metal-Binding Site in the Catalytic Subunit of Anaerobic Ribonucleotide Reductase.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8BT3
 
 | |
8BT4
 
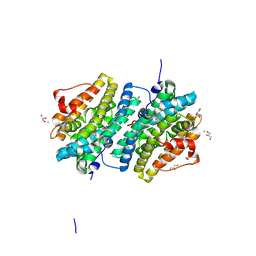 | | Ribonucleotide Reductase class Ie R2 from Mesoplasma florum, radical-lost ground state | | Descriptor: | CALCIUM ION, GLYCEROL, Ribonucleoside-diphosphate reductase | | Authors: | Lebrette, H, Srinivas, V, Hogbom, M. | | Deposit date: | 2022-11-27 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of a ribonucleotide reductase R2 protein radical.
Science, 382, 2023
|
|
6ZJK
 
 | |
3DHZ
 
 | |
5IM3
 
 | |
4R1R
 
 | |
3R1R
 
 | |
7Q3C
 
 | |
2R1R
 
 | |
7AIL
 
 | |
7AGJ
 
 | | Ribonucleotide Reductase R1 protein from Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Rehling, D, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2020-09-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Investigation of Class I Ribonucleotide Reductase from the Hyperthermophile Aquifex aeolicus.
Biochemistry, 61, 2022
|
|
7AIK
 
 | | Ribonucleotide Reductase R2 protein from Aquifex aeolicus | | Descriptor: | FE (II) ION, Ribonucleoside-diphosphate reductase subunit beta,Ribonucleoside-diphosphate reductase subunit beta | | Authors: | Rehling, D, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2020-09-27 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Investigation of Class I Ribonucleotide Reductase from the Hyperthermophile Aquifex aeolicus.
Biochemistry, 61, 2022
|
|
7P37
 
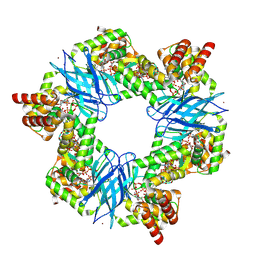 | | Streptomyces coelicolor ATP-loaded NrdR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ZINC ION | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
7P3Q
 
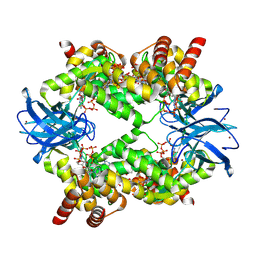 | | Streptomyces coelicolor dATP/ATP-loaded NrdR octamer | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ... | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
7P3F
 
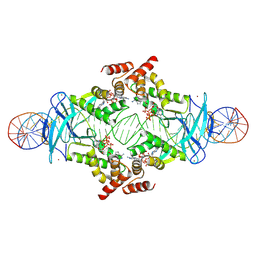 | |
7Q39
 
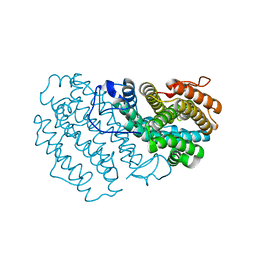 | |
1RNR
 
 | |
2XAZ
 
 | |
2XAX
 
 | |
2XO5
 
 | | RIBONUCLEOTIDE REDUCTASE Y731NH2Y MODIFIED R1 SUBUNIT OF E. COLI | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Minnihan, E.C, Seyedsayamdost, M.R, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-08-09 | | Release date: | 2010-08-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Kinetics of Radical Intermediate Formation and Deoxynucleotide Production in 3-Aminotyrosine- Substituted Escherichia Coli Ribonucleotide Reductases.
J.Am.Chem.Soc., 133, 2011
|
|
