5KVA
 
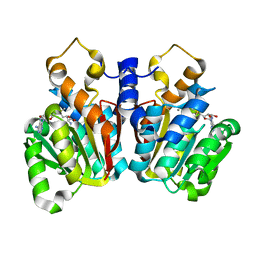 | | Crystal Structure of sorghum caffeoyl-CoA O-methyltransferase (CCoAOMT) | | Descriptor: | CALCIUM ION, S-ADENOSYLMETHIONINE, caffeoyl-CoA O-methyltransferase | | Authors: | Walker, A.M, Sattler, S.A, Regner, M, Jones, J.P, Ralph, J, Vermerris, W, Sattler, S.E, Kang, C. | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | The Structure and Catalytic Mechanism of Sorghum bicolor Caffeoyl-CoA O-Methyltransferase.
Plant Physiol., 172, 2016
|
|
1JL6
 
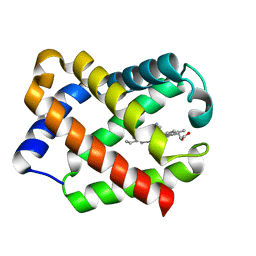 | | Crystal Structure of CN-Ligated Component IV Glycera Dibranchiata Monomeric Hemoglobin | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, monomer hemoglobin component IV | | Authors: | Park, H.J, Yang, C, Treff, N, Satterlee, J.D, Kang, C. | | Deposit date: | 2001-07-16 | | Release date: | 2002-07-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Unligated and CN-Ligated Glycera dibranchiata Monomer Ferric Hemoglobin Components III and IV
Proteins, 49, 2002
|
|
4RNS
 
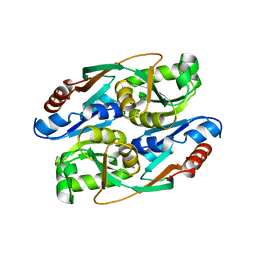 | | PcpR inducer binding domain (apo-form) | | Descriptor: | PCP degradation transcriptional activation protein | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-25 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
4RPO
 
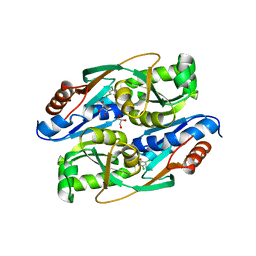 | | PcpR inducer binding domain (Complex with 2,4,6-trichlorophenol) | | Descriptor: | 2,4,6-trichlorophenol, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
4RPN
 
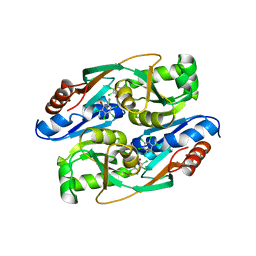 | | PcpR inducer binding domain complex with pentachlorophenol | | Descriptor: | PCP degradation transcriptional activation protein, PENTACHLOROPHENOL | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
6VBY
 
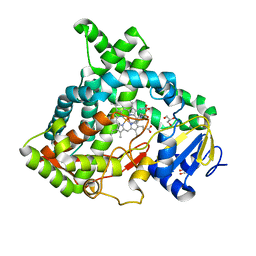 | | Cinnamate 4-hydroxylase (C4H1) from Sorghum bicolor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cinnamic acid 4-hydroxylase, GLYCEROL, ... | | Authors: | Zhang, B, Kang, C, Lewis, K.M. | | Deposit date: | 2019-12-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function of the Cytochrome P450 Monooxygenase Cinnamate 4-hydroxylase fromSorghum bicolor.
Plant Physiol., 183, 2020
|
|
1S3O
 
 | |
6WM6
 
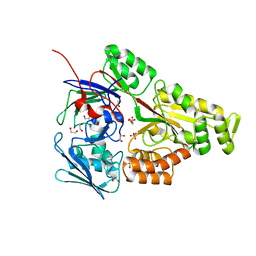 | | Periplasmic EDTA-binding protein EppA, tetragonal | | Descriptor: | 1,2-ETHANEDIOL, Extracellular solute-binding protein, family 5, ... | | Authors: | Lewis, K.M, Sattler, S.A, Greene, C.L, Xun, L, Kang, C. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Structural Basis of the Binding of Various Aminopolycarboxylates by the Periplasmic EDTA-Binding Protein EppA from Chelativorans sp. BNC1.
Int J Mol Sci, 21, 2020
|
|
6WM7
 
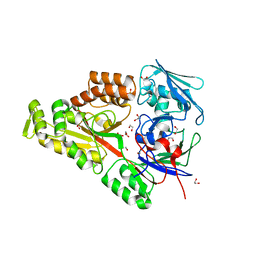 | | Periplasmic EDTA-binding protein EppA, orthorhombic | | Descriptor: | 1,2-ETHANEDIOL, Extracellular solute-binding protein, family 5, ... | | Authors: | Lewis, K.M, Greene, C.L, Sattler, S.A, Xun, L, Kang, C. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The Structural Basis of the Binding of Various Aminopolycarboxylates by the Periplasmic EDTA-Binding Protein EppA from Chelativorans sp. BNC1.
Int J Mol Sci, 21, 2020
|
|
6XND
 
 | | Avidin-Biotin-Phenol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin, N-[2-(2-hydroxy-5-nitrophenyl)ethyl]-5-[(3aS,4S,6aS)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide | | Authors: | Ahmadvand, P, Kang, C. | | Deposit date: | 2020-07-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Ligand-Directed Nitrophenol Carbonate for Transient in situ Bioconjugation and Drug Delivery
Chemmedchem, 15, 2020
|
|
2CF5
 
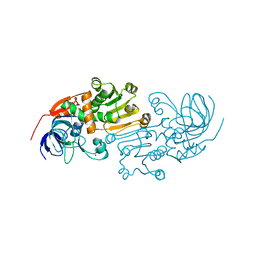 | | Crystal Structures of the Arabidopsis Cinnamyl Alcohol Dehydrogenases, AtCAD5 | | Descriptor: | CINNAMYL ALCOHOL DEHYDROGENASE, ZINC ION | | Authors: | Youn, B, Camacho, R, Moinuddin, S, Lee, C, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-02-16 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures and Catalytic Mechanisms of the Arabidopsis Cinnamyl Alcohol Dehydrogenases Atcad5 and Atcad4
Org.Biomol.Chem., 4, 2006
|
|
2J3H
 
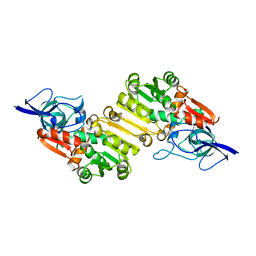 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Apo form | | Descriptor: | NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
2CF6
 
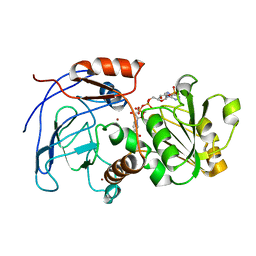 | | Crystal Structures of the Arabidopsis Cinnamyl Alcohol Dehydrogenases AtCAD5 | | Descriptor: | CINNAMYL ALCOHOL DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Youn, B, Camacho, R, Moinuddin, S.G, Lee, C, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-02-16 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures and Catalytic Mechanisms of the Arabidopsis Cinnamyl Alcohol Dehydrogenases Atcad5 and Atcad4
Org.Biomol.Chem., 4, 2006
|
|
2J3K
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex II | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
2J3J
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex I | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
7SUX
 
 | |
7SUZ
 
 | |
7SV0
 
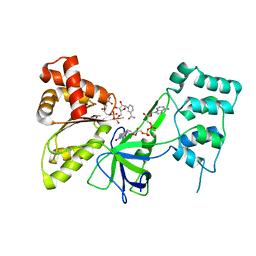 | |
2J3I
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Binary Complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
8DJR
 
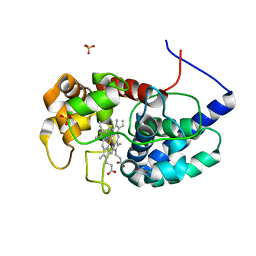 | | Cytosolic ascorbate peroxidase from Sorghum bicolor | | Descriptor: | L-ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids.
Plant Physiol., 192, 2023
|
|
8DJX
 
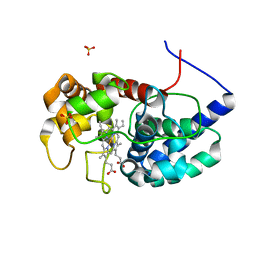 | | Cytosolic ascorbate peroxidase from Sorghum bicolor - Compound II | | Descriptor: | L-ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids.
Plant Physiol., 192, 2023
|
|
8DJS
 
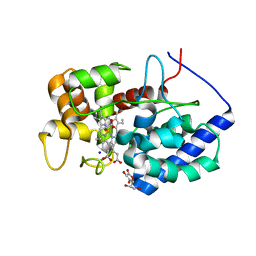 | |
8DJU
 
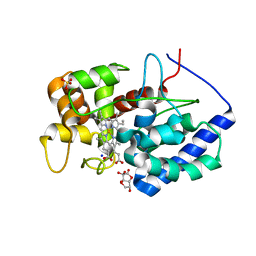 | | Cytosolic ascorbate peroxidase from Sorghum bicolor - bicyclic dehydroascorbic acid complex | | Descriptor: | (3aS,6S,6aR)-3,3,3a,6-tetrahydroxytetrahydrofuro[3,2-b]furan-2(3H)-one (non-preferred name), L-ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids.
Plant Physiol., 192, 2023
|
|
8DJT
 
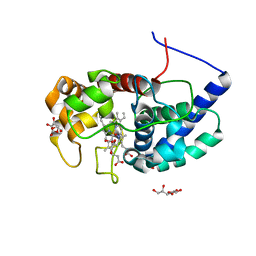 | |
8DJW
 
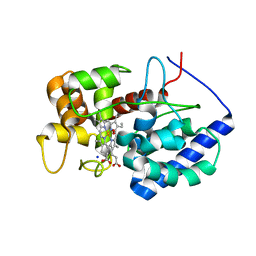 | |
