5HI8
 
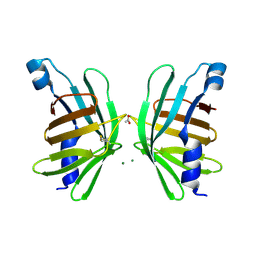 | | Structure of T-type Phycobiliprotein Lyase CpeT from Prochlorococcus phage P-HM1 | | Descriptor: | ACETATE ION, Antenna protein, MAGNESIUM ION | | Authors: | Gasper, R, Schwach, J, Frankenberg-Dinkel, N, Hofmann, E. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct Features of Cyanophage-encoded T-type Phycobiliprotein Lyase Phi CpeT: THE ROLE OF AUXILIARY METABOLIC GENES.
J. Biol. Chem., 292, 2017
|
|
3IIS
 
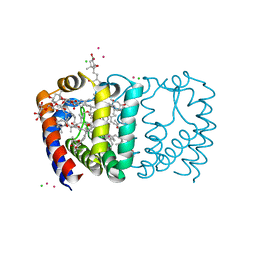 | | Structure of the reconstituted Peridinin-Chlorophyll a-Protein (RFPCP) | | Descriptor: | (2S)-3-[(6-O-alpha-D-galactopyranosyl-beta-D-galactopyranosyl)oxy]-2-[(3Z,6Z,9Z,12Z,15Z)-octadeca-3,6,9,12,15-pentaenoyloxy]propyl (5Z,8Z,11Z,14Z,17Z)-icosa-5,8,11,14,17-pentaenoate, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Schulte, T, Hofmann, E. | | Deposit date: | 2009-08-03 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of a single peridinin sensing Chl-a excitation in reconstituted PCP by crystallography and spectroscopy.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5LAL
 
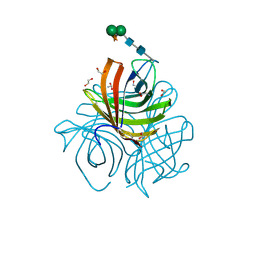 | | Structure of Arabidopsis dirigent protein AtDIR6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dirigent protein 6, ... | | Authors: | Gasper, R, Kolesinski, P, Terlecka, B, Effenberger, I, Schaller, A, Hofmann, E. | | Deposit date: | 2016-06-14 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dirigent Protein Mode of Action Revealed by the Crystal Structure of AtDIR6.
Plant Physiol., 172, 2016
|
|
5LA3
 
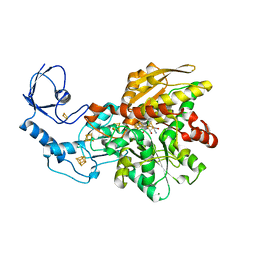 | | [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant E279A | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, Iron hydrogenase 1, ... | | Authors: | Duan, J, Esselborn, J, Hofmann, E, Winkler, M, Happe, T. | | Deposit date: | 2016-06-13 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Accumulating the hydride state in the catalytic cycle of [FeFe]-hydrogenases.
Nat Commun, 8, 2017
|
|
4AK9
 
 | |
3IIU
 
 | | Structure of the reconstituted Peridinin-Chlorophyll a-Protein (RFPCP) mutant N89L | | Descriptor: | (2S)-3-[(6-O-alpha-D-galactopyranosyl-beta-D-galactopyranosyl)oxy]-2-[(3Z,6Z,9Z,12Z,15Z)-octadeca-3,6,9,12,15-pentaenoyloxy]propyl (5Z,8Z,11Z,14Z,17Z)-icosa-5,8,11,14,17-pentaenoate, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Schulte, T, Hofmann, E. | | Deposit date: | 2009-08-03 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of a single peridinin sensing Chl-a excitation in reconstituted PCP by crystallography and spectroscopy.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5IXB
 
 | | Structure of human Melanoma Inhibitory Activity (MIA) Protein in complex with Pyrimidin-2-amine | | Descriptor: | ACETATE ION, Melanoma-derived growth regulatory protein, PYRIMIDIN-2-AMINE | | Authors: | Yip, K.T, Gasper, R, Zhong, X.Y, Seibel, N, Puetz, S, Autzen, J, Scherkenbeck, J, Hofmann, E, Stoll, R. | | Deposit date: | 2016-03-23 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Small Molecules Antagonise the MIA-Fibronectin Interaction in Malignant Melanoma.
Sci Rep, 6, 2016
|
|
4CQ6
 
 | | The crystal structure of the allene oxide cyclase 2 from Arabidopsis thaliana with bound inhibitor - vernolic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (9Z)-11-[(2R,3S)-3-pentyloxiran-2-yl]undec-9-enoic acid, ALLENE OXIDE CYCLASE 2, ... | | Authors: | Terlecka, B.A, Pollmann, S, Hofmann, E. | | Deposit date: | 2014-02-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Ligand-Bound Structures of the Aoc2 from A. Thaliana and the Implications for the Catalytic Mechanism
To be Published
|
|
4CQ7
 
 | | The crystal structure of the allene oxide cyclase 2 from Arabidopsis thaliana with bound product - OPDA | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 9S,13R-12-OXOPHYTODIENOIC ACID, ALLENE OXIDE CYCLASE 2, ... | | Authors: | Terlecka, B.A, Pollmann, S, Hofmann, E. | | Deposit date: | 2014-02-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Ligand-Bound Structures of the Aoc2 from A. Thaliana and the Implications for the Catalytic Mechanism
To be Published
|
|
2WJL
 
 | | Bacteriorhodopsin mutant E194D | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Potschies, M, Wolf, S, Freier, E, Hofmann, E, Gerwert, K. | | Deposit date: | 2009-05-27 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Directional proton transfer in membrane proteins achieved through protonated protein-bound water molecules: a proton diode.
Angew. Chem. Int. Ed. Engl., 49, 2010
|
|
2WJK
 
 | | Bacteriorhodopsin mutant E204D | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Potschies, M, Hofmann, E, Gerwert, K. | | Deposit date: | 2009-05-27 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Directional proton transfer in membrane proteins achieved through protonated protein-bound water molecules: a proton diode.
Angew. Chem. Int. Ed. Engl., 49, 2010
|
|
2YMY
 
 | | Structure of the murine Nore1-Sarah domain | | Descriptor: | CADMIUM ION, RAS ASSOCIATION DOMAIN-CONTAINING PROTEIN 5 | | Authors: | Makbul, C, Schwarz, D, Aruxandei, D.C, Wolf, E, Hofmann, E, Herrmann, C. | | Deposit date: | 2012-10-10 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and Thermodynamic Characterization of Nore1-Sarah: A Small, Helical Module Important in Signal Transduction Networks.
Biochemistry, 52, 2013
|
|
2GMR
 
 | | Photosynthetic reaction center mutant from Rhodobacter sphaeroides with Asp L210 replaced with Asn | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Stachnik, J.M, Hermes, S, Gerwert, K, Hofmann, E. | | Deposit date: | 2006-04-07 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Proton uptake in the reaction center mutant L210DN from Rhodobacter sphaeroides via protonated water molecules.
Biochemistry, 45, 2006
|
|
5BYQ
 
 | | Semisynthetic [FeFe]-hydrogenase CpI with oxodithiolato-bridged [2Fe] cofactor | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Esselborn, J, Muraki, N, Engelbrecht, V, Hofmann, E, Kurisu, G, Happe, T. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A structural view of synthetic cofactor integration into [FeFe]-hydrogenases.
Chem Sci, 7, 2016
|
|
5BYS
 
 | | Semisynthetic [FeFe]-hydrogenase CpI with sulfur-dithiolato-bridged [2Fe] cofactor | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, Iron hydrogenase 1, ... | | Authors: | Esselborn, J, Muraki, N, Engelbrecht, V, Hofmann, E, Kurisu, G, Happe, T. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A structural view of synthetic cofactor integration into [FeFe]-hydrogenases.
Chem Sci, 7, 2016
|
|
5BYR
 
 | | Semisynthetic [FeFe]-hydrogenase CpI with propane-dithiolato-bridged [2Fe] cofactor | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Esselborn, J, Muraki, N, Engelbrecht, V, Hofmann, E, Kurisu, G, Happe, T. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A structural view of synthetic cofactor integration into [FeFe]-hydrogenases.
Chem Sci, 7, 2016
|
|
2VCL
 
 | |
2VCK
 
 | |
2VGR
 
 | |
2X9J
 
 | |
2X9O
 
 | | STRUCTURE OF 15, 16- DIHYDROBILIVERDIN:FERREDOXIN OXIDOREDUCTASE (PebA) | | Descriptor: | 15,16-DIHYDROBILIVERDIN-FERREDOXIN OXIDOREDUCTASE, BILIVERDINE IX ALPHA | | Authors: | Busch, A.W.U, Frankenberg-Dinkel, N, Hofmann, E. | | Deposit date: | 2010-03-23 | | Release date: | 2011-03-30 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Mechanistic Insight Into the Ferredoxin-Mediated Two-Electron Reduction of Bilins.
Biochem.J., 439, 2011
|
|
2X1Z
 
 | | Structure of Peridinin-Chlorophyll-Protein reconstituted with Chl-d | | Descriptor: | CADMIUM ION, CHLORIDE ION, CHLOROPHYLL D, ... | | Authors: | Schulte, T, Hiller, R.G, Hofmann, E. | | Deposit date: | 2010-01-09 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures of the Peridinin-Chlorophyll-Protein Reconstituted with Different Chlorophylls.
FEBS Lett., 584, 2010
|
|
1P0Z
 
 | | Sensor Kinase CitA binding domain | | Descriptor: | CITRATE ANION, MO(VI)(=O)(OH)2 CLUSTER, SODIUM ION, ... | | Authors: | Reinelt, S, Hofmann, E, Gerharz, T, Bott, M, Madden, D.R. | | Deposit date: | 2003-04-11 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain.
J.Biol.Chem., 278, 2003
|
|
8PVM
 
 | | formaldehyde-inhibited [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant C299D | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FORMYL GROUP, ... | | Authors: | Duan, J, Hofmann, E, Happe, T. | | Deposit date: | 2023-07-18 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Insights into the Molecular Mechanism of Formaldehyde Inhibition of [FeFe]-Hydrogenases.
J.Am.Chem.Soc., 145, 2023
|
|
8QM3
 
 | |
