1MKC
 
 | | C-TERMINAL DOMAIN OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
1MKN
 
 | | N-TERMINAL HALF OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
1UFM
 
 | | Solution structure of the PCI domain | | Descriptor: | COP9 complex subunit 4 | | Authors: | Suzuki, S, Hatanaka, H, Kigawa, T, Shirouzu, M, Hayashizaki, Y, The RIKEN Genome Exploration Research Group Phase I & II Teams, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-02 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PCI domain
To be Published
|
|
1UHF
 
 | | Solution Structure of the third SH3 domain of human intersectin 2(KIAA1256) | | Descriptor: | INTERSECTIN 2 | | Authors: | Suzuki, S, Hatanaka, H, Koshiba, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-03 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the third SH3 domain of human intersectin 2(KIAA1256)
To be Published
|
|
1UHU
 
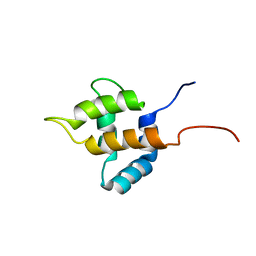 | | Solution structure of the retroviral Gag MA-like domain of RIKEN cDNA 3110009E22 | | Descriptor: | product of RIKEN cDNA 3110009E22 | | Authors: | Suzuki, S, Hatanaka, H, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the retroviral Gag MA-like domain of RIKEN cDNA 3110009E22
To be Published
|
|
1AHM
 
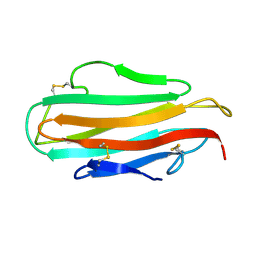 | | DER F 2, THE MAJOR MITE ALLERGEN FROM DERMATOPHAGOIDES FARINAE, NMR, 10 STRUCTURES | | Descriptor: | DER F 2 | | Authors: | Ichikawa, S, Hatanaka, H, Yuuki, T, Iwamoto, N, Ogura, K, Okumura, Y, Inagaki, F. | | Deposit date: | 1997-04-07 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Der f 2, the major mite allergen for atopic diseases.
J.Biol.Chem., 273, 1998
|
|
1AHK
 
 | | DER F 2, THE MAJOR MITE ALLERGEN FROM DERMATOPHAGOIDES FARINAE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DER F 2 | | Authors: | Ichikawa, S, Hatanaka, H, Yuuki, T, Iwamoto, N, Ogura, K, Okumura, Y, Inagaki, F. | | Deposit date: | 1997-04-07 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Der f 2, the major mite allergen for atopic diseases.
J.Biol.Chem., 273, 1998
|
|
1D4M
 
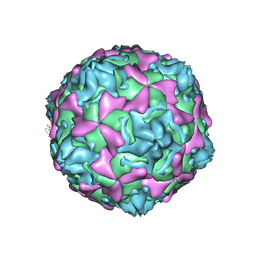 | | THE CRYSTAL STRUCTURE OF COXSACKIEVIRUS A9 TO 2.9 A RESOLUTION | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, PROTEIN (COXSACKIEVIRUS A9) | | Authors: | Hendry, E, Hatanaka, H, Fry, E, Smyth, M, Tate, J, Stanway, G, Santti, J, Maaronen, M, Hyypia, T, Stuart, D. | | Deposit date: | 1999-10-04 | | Release date: | 1999-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses.
Structure Fold.Des., 7, 1999
|
|
1HSR
 
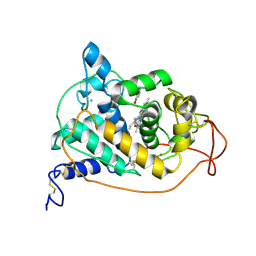 | | BINDING MODE OF BENZHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1997-07-01 | | Release date: | 1998-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding mode of benzhydroxamic acid to Arthromyces ramosus peroxidase shown by X-ray crystallographic analysis of the complex at 1.6 A resolution.
FEBS Lett., 412, 1997
|
|
1TCH
 
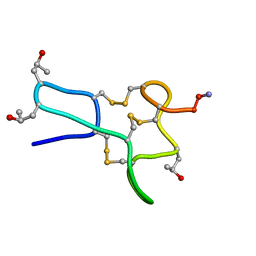 | |
1TCG
 
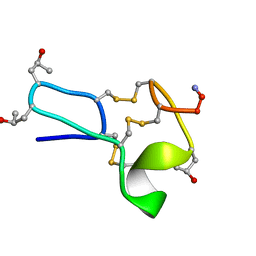 | |
1TCK
 
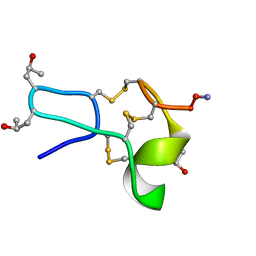 | |
1TCJ
 
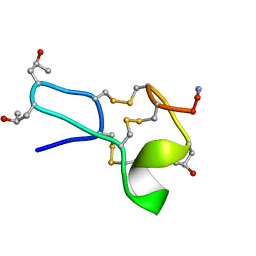 | |
1QG1
 
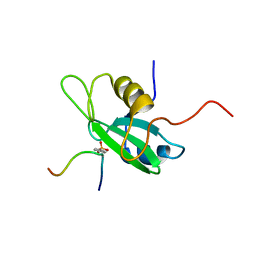 | |
1ARP
 
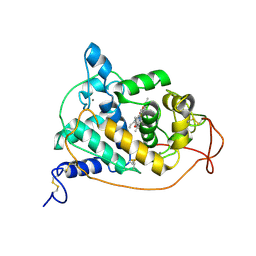 | |
1CK6
 
 | | BINDING MODE OF SALICYLHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN (PEROXIDASE), ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1999-04-28 | | Release date: | 1999-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of salicylhydroxamic acid and several aromatic donor molecules to Arthromyces ramosus peroxidase, investigated by X-ray crystallography, optical difference spectroscopy, NMR relaxation, molecular dynamics, and kinetics.
Biochemistry, 38, 1999
|
|
1GZA
 
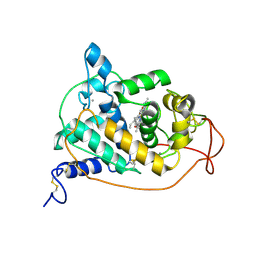 | | PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Binding of iodide to Arthromyces ramosus peroxidase investigated with X-ray crystallographic analysis, 1H and 127I NMR spectroscopy, and steady-state kinetics.
J.Biol.Chem., 272, 1997
|
|
2E3A
 
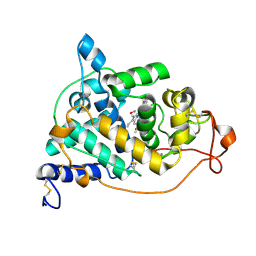 | | Crystal structure of the NO-bound form of Arthromyces ramosus peroxidase at 1.3 Angstroms resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NITRIC OXIDE, ... | | Authors: | Fukuyama, K, Okada, T. | | Deposit date: | 2006-11-22 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of cyanide, nitric oxide and hydroxylamine complexes of Arthromyces ramosusperoxidase at 100 K refined to 1.3 A resolution: coordination geometries of the ligands to the haem iron
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2E39
 
 | | Crystal structure of the CN-bound form of Arthromyces ramosus peroxidase at 1.3 Angstroms resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Okada, T. | | Deposit date: | 2006-11-22 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of cyanide, nitric oxide and hydroxylamine complexes of Arthromyces ramosusperoxidase at 100 K refined to 1.3 A resolution: coordination geometries of the ligands to the haem iron
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2E3B
 
 | | Crystal structure of the HA-bound form of Arthromyces ramosus peroxidase at 1.3 Angstroms resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HYDROXYAMINE, ... | | Authors: | Fukuyama, K, Okada, T. | | Deposit date: | 2006-11-22 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of cyanide, nitric oxide and hydroxylamine complexes of Arthromyces ramosusperoxidase at 100 K refined to 1.3 A resolution: coordination geometries of the ligands to the haem iron
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
1C8I
 
 | | BINDING MODE OF HYDROXYLAMINE TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HYDROXYAMINE, ... | | Authors: | Wariishi, H, Nonaka, D, Johjima, T, Nakamura, N, Naruta, Y, Kubo, K, Fukuyama, K. | | Deposit date: | 2000-05-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct binding of hydroxylamine to the heme iron of Arthromyces ramosus peroxidase. Substrate analogue that inhibits compound I formation in a competetive manner.
J.Biol.Chem., 275, 2000
|
|
1GZB
 
 | | PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PEROXIDASE, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pentacoordination of the heme iron of Arthromyces ramosus peroxidase shown by a 1.8 A resolution crystallographic study at pH 4.5.
FEBS Lett., 378, 1996
|
|
1GIB
 
 | | MU-CONOTOXIN GIIIB, NMR | | Descriptor: | MU-CONOTOXIN GIIIB | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-04-17 | | Release date: | 1996-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mu-conotoxin GIIIB, a specific blocker of skeletal muscle sodium channels.
Biochemistry, 35, 1996
|
|
1ARX
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARY
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
