8CIO
 
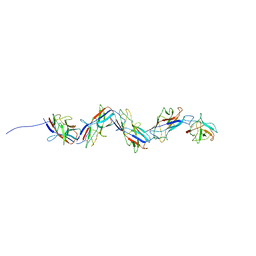 | |
3CWO
 
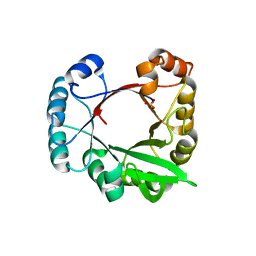 | | A beta/alpha-barrel built by the combination of fragments from different folds | | Descriptor: | SULFATE ION, beta/alpha-barrel protein based on 1THF and 1TMY | | Authors: | Bharat, T.A.M, Eisenbeis, S, Zeth, K, Hocker, B. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A beta alpha-barrel built by the combination of fragments from different folds.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5N97
 
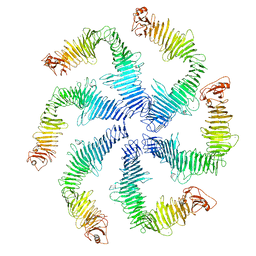 | | Structure of the C. crescentus S-layer | | Descriptor: | CALCIUM ION, S-layer protein rsaA | | Authors: | Bharat, T.A, Hagen, W.J, Briggs, J.A, Lowe, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the hexagonal surface layer on Caulobacter crescentus cells.
Nat Microbiol, 2, 2017
|
|
8BQE
 
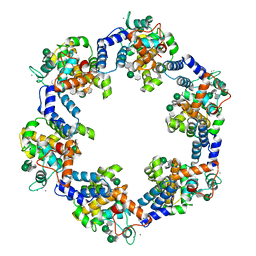 | | In situ structure of the Caulobacter crescentus S-layer | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose, CALCIUM ION, S-layer protein rsaA | | Authors: | von Kuegelgen, A, Bharat, T. | | Deposit date: | 2022-11-21 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0.
Elife, 11, 2022
|
|
8C8K
 
 | |
8C8N
 
 | |
8C8M
 
 | |
8C8L
 
 | |
8C8O
 
 | |
5O01
 
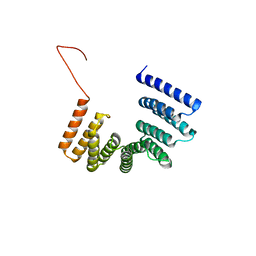 | | Crystal structure of BtubC (BKLC) from P. vanneervenii | | Descriptor: | BKLC (bacterial kinesin-light chain-like) | | Authors: | Lowe, J, Deng, X. | | Deposit date: | 2017-05-15 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Four-stranded mini microtubules formed by Prosthecobacter BtubAB show dynamic instability.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8C8R
 
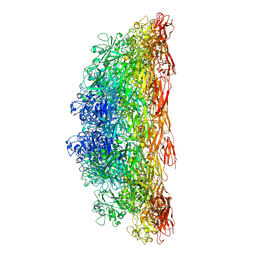 | |
8CKA
 
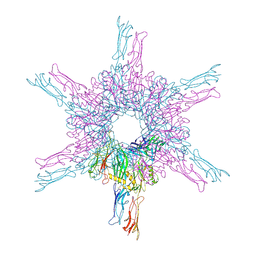 | | Deinococcus radidurans HPI S-layer | | Descriptor: | Hexagonally packed intermediate-layer surface protein | | Authors: | von Kuegelgen, A, Yamashita, K, Murshudov, G, Bharat, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Interdigitated immunoglobulin arrays form the hyperstable surface layer of the extremophilic bacterium Deinococcus radiodurans.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BSH
 
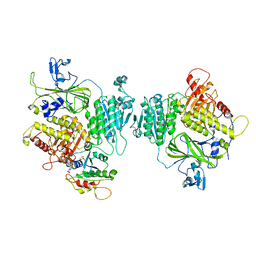 | | COPII inner coat | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein transport protein SEC23, ... | | Authors: | Zanetti, G, Pyle, E.W. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0.
Elife, 11, 2022
|
|
