8BPR
 
 | | Complex of RecF-RecO-RecR-DNA from Thermus thermophilus (low resolution reconstruction). | | Descriptor: | DNA repair protein RecO, DNA replication and repair protein RecF, MAGNESIUM ION, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5M7X
 
 | | Translation initiation factor 4E in complex with (RP)-m2(7,2'O)GppSepG mRNA 5' cap analog (beta-Se-ARCA D1) | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(selanyl)phosphoryl] hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Translation initiation factor 4E in complex with (RP)-m2(7,2'O)GppSepG mRNA 5' cap analog (beta-Se-ARCA D1)
To Be Published
|
|
5M7W
 
 | | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D2) | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(sulfanyl)phosphoryl] hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D2)
To Be Published
|
|
5M7V
 
 | | Translation initiation factor 4E in complex with (RP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D1) | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(sulfanyl)phosphoryl] hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Translation initiation factor 4E in complex with (RP)-m2(7,2'O)GppSpG mRNA 5' cap analog (beta-S-ARCA D1)
To Be Published
|
|
5M84
 
 | | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpA mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-sulfanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanyl-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpA mRNA 5' cap analog
To Be Published
|
|
5M7Z
 
 | | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSepG mRNA 5' cap analog (beta-Se-ARCA D2) | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 4E, GLYCEROL, ... | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSepG mRNA 5' cap analog (beta-Se-ARCA D2)
To Be Published
|
|
8BDC
 
 | | Human apo TRPM8 in a closed state (composite map) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Palchevskyi, S, Czarnocki-Cieciura, M, Vistoli, G, Gervasoni, S, Nowak, E, Beccari, A.R, Nowotny, M, Talarico, C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structure of human TRPM8 channel.
Commun Biol, 6, 2023
|
|
7R08
 
 | | Abortive infection DNA polymerase Abi-P2 | | Descriptor: | Reverse transcriptase | | Authors: | Gapinska, M.A, Figiel, M, Czarnocki Cieciura, M, Nowotny, M, Zajko, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
7R06
 
 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis | | Descriptor: | AbiK, DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3') | | Authors: | Figiel, M, Nowotny, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
7R07
 
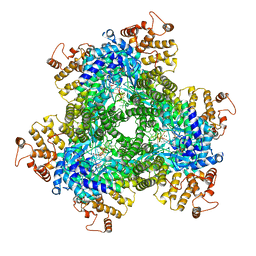 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis | | Descriptor: | AbiK, DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), MAGNESIUM ION | | Authors: | Figiel, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W, Nowotny, M. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
6YWN
 
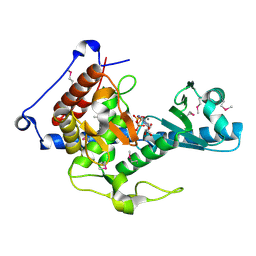 | | CutA in complex with CMPCPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, CALCIUM ION, CutA | | Authors: | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-05 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
6YWP
 
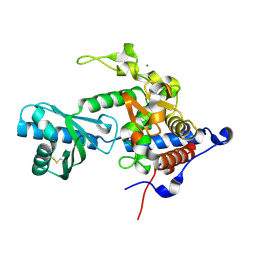 | | Structure of apo-CutA | | Descriptor: | CutA, MAGNESIUM ION | | Authors: | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-05 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
6F4A
 
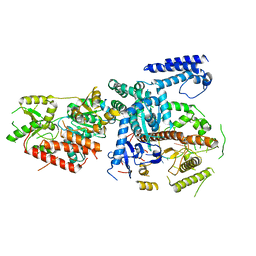 | | Yeast mitochondrial RNA degradosome complex mtEXO | | Descriptor: | Exoribonuclease II, mitochondrial, RNA (5'-R(P*AP*GP*AP*UP*AP*C)-3'), ... | | Authors: | Razew, M, Nowak, E, Nowotny, M. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components.
Nat Commun, 9, 2018
|
|
6F3H
 
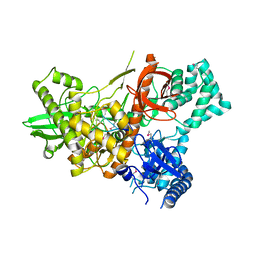 | | Crystal structure of Dss1 exoribonuclease active site mutant D477N from Candida glabrata | | Descriptor: | Exoribonuclease II, mitochondrial, MAGNESIUM ION, ... | | Authors: | Razew, M, Nowak, E, Nowotny, M. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components.
Nat Commun, 9, 2018
|
|
1RL1
 
 | | Solution structure of human Sgt1 CS domain | | Descriptor: | Suppressor of G2 allele of SKP1 homolog | | Authors: | Lee, Y.-T, Jacob, J, Michowski, W, Nowotny, M, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2003-11-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Human Sgt1 Binds HSP90 through the CHORD-Sgt1 Domain and Not the Tetratricopeptide Repeat Domain
J.Biol.Chem., 279, 2004
|
|
4XM5
 
 | | C. glabrata Slx1. | | Descriptor: | CHLORIDE ION, Structure-specific endonuclease subunit SLX1, ZINC ION | | Authors: | Gaur, V, Wyatt, H.D.M, Komorowska, W, Szczepanowski, R.H, de Sanctis, D, Gorecka, K.M, West, S.C, Nowotny, M. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and Mechanistic Analysis of the Slx1-Slx4 Endonuclease.
Cell Rep, 10, 2015
|
|
4LD0
 
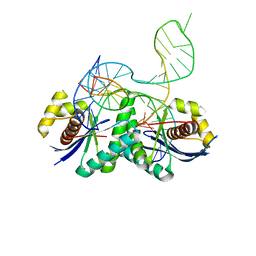 | | T. thermophilus RuvC in complex with Holliday junction substrate | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC, DNA 11-MER, DNA 13-MER, ... | | Authors: | Gorecka, K.M, Komorowska, W, Nowotny, M. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Crystal structure of RuvC resolvase in complex with Holliday junction substrate.
Nucleic Acids Res., 41, 2013
|
|
7O0H
 
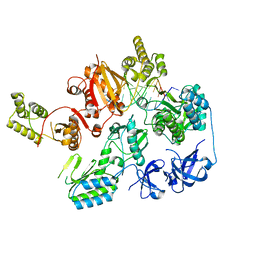 | |
7O0G
 
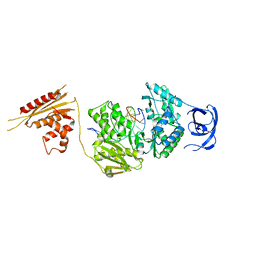 | |
4HHT
 
 | |
7Z0Z
 
 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis, Y44F variant | | Descriptor: | AbiK | | Authors: | Figiel, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W, Nowotny, M. | | Deposit date: | 2022-02-24 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
6YWO
 
 | | CutA in complex with A3 RNA | | Descriptor: | CHLORIDE ION, CutA, MAGNESIUM ION, ... | | Authors: | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-05 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
8OZ7
 
 | |
8R42
 
 | | Structure of CHI3L1 in complex with inhibititor 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2~{R})-2-[(4-chlorophenyl)methyl]pyrrolidin-1-yl]piperidin-1-yl]pyridine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | Deposit date: | 2023-11-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
6S16
 
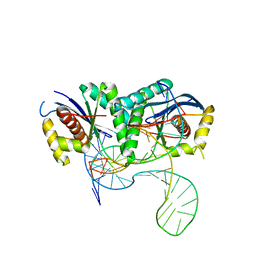 | | T. thermophilus RuvC in complex with Holliday junction substrate | | Descriptor: | CHLORIDE ION, Crossover junction endodeoxyribonuclease RuvC, DNA (33-MER), ... | | Authors: | Gorecka, K.M, Krepl, M, Szlachcic, A, Poznanski, J, Sponer, J, Nowotny, M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.409 Å) | | Cite: | RuvC uses dynamic probing of the Holliday junction to achieve sequence specificity and efficient resolution.
Nat Commun, 10, 2019
|
|
